7ZZ1
 
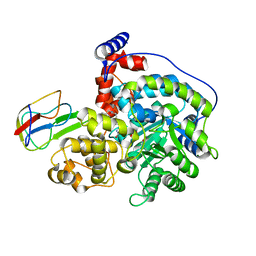 | | Cryo-EM structure of "CT react" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | BIOTIN, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ4
 
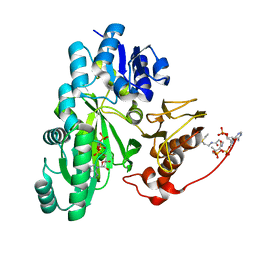 | | Cryo-EM structure of "BC closed" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ0
 
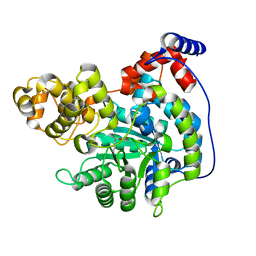 | | Cryo-EM structure of "CT empty" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Pyruvate carboxylase | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZYY
 
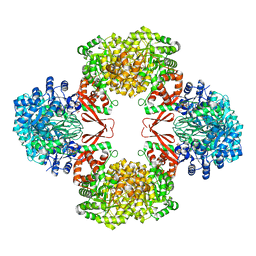 | | Cryo-EM structure of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, BICARBONATE ION, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ6
 
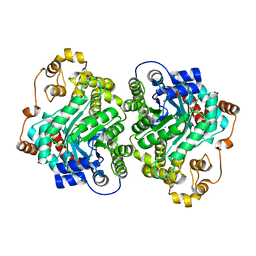 | | Cryo-EM structure of "CT-CT dimer" of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, PYRUVIC ACID, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ5
 
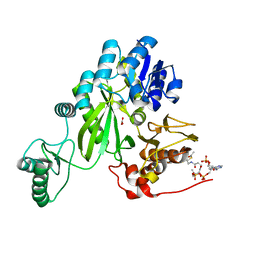 | | Cryo-EM structure of "BC open" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, BICARBONATE ION, Pyruvate carboxylase | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ3
 
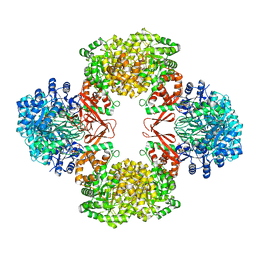 | | Cryo-EM structure of "BC react" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZYZ
 
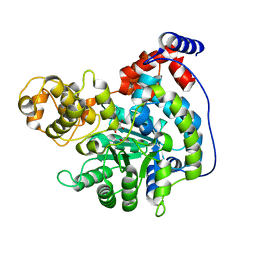 | | Cryo-EM structure of "CT oxa" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | MANGANESE (II) ION, OXALOACETATE ION, Pyruvate carboxylase | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ8
 
 | | Cryo-EM structure of Lactococcus lactis pyruvate carboxylase with acetyl-CoA and cyclic di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7PDC
 
 | |
6FO1
 
 | | Human R2TP subcomplex containing 1 RUVBL1-RUVBL2 hexamer bound to 1 RBD domain from RPAP3. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RNA polymerase II-associated protein 3, RuvB-like 1, ... | | Authors: | Martino, F, Munoz-Hernandez, H, Rodriguez, C.F, Pearl, L.H, Llorca, O. | | Deposit date: | 2018-02-05 | | Release date: | 2018-04-04 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | RPAP3 provides a flexible scaffold for coupling HSP90 to the human R2TP co-chaperone complex.
Nat Commun, 9, 2018
|
|
6EHT
 
 | | Modulation of PCNA sliding surface by p15PAF suggests a suppressive mechanism for cisplatin-induced DNA lesion bypass by pol eta holoenzyme | | Descriptor: | DNA (5'-D(P*AP*TP*AP*CP*GP*AP*TP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*AP*TP*CP*GP*TP*AP*T)-3'), PCNA-associated factor, ... | | Authors: | De March, M, Barrera-Vilarmau, S, Mentegari, E, Merino, N, Bressan, E, Maga, G, Crehuet, R, Onesti, S, Blanco, F.J, De Biasio, A. | | Deposit date: | 2017-09-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | p15PAF binding to PCNA modulates the DNA sliding surface.
Nucleic Acids Res., 46, 2018
|
|
9G1V
 
 | | Yeast RNA polymerase I elongation complex stalled by an apurinic site | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Santos-Aledo, A, Plaza-Pegueroles, A, Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM uncovers a sequential mechanism for RNA polymerase I pausing and stalling at abasic DNA lesions.
Nat Commun, 16, 2025
|
|
9G26
 
 | | Yeast RNA polymerase I elongation complex stalled by an apurinic site, closed state | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Santos-Aledo, A, Plaza-Pegueroles, A, Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM uncovers a sequential mechanism for RNA polymerase I pausing and stalling at abasic DNA lesions.
Nat Commun, 16, 2025
|
|
9G27
 
 | | Yeast RNA polymerase I elongation complex stalled by an apurinic site, pre-translocation state | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Santos-Aledo, A, Plaza-Pegueroles, A, Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM uncovers a sequential mechanism for RNA polymerase I pausing and stalling at abasic DNA lesions.
Nat Commun, 16, 2025
|
|
9G1X
 
 | | Yeast RNA polymerase I elongation complex stalled by an apurinic site, 11-subunit | | Descriptor: | DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, DNA-directed RNA polymerase I subunit RPA190, ... | | Authors: | Santos-Aledo, A, Plaza-Pegueroles, A, Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM uncovers a sequential mechanism for RNA polymerase I pausing and stalling at abasic DNA lesions.
Nat Commun, 16, 2025
|
|
9G23
 
 | | Yeast RNA polymerase I elongation complex stalled by an apurinic site bound to nucleotide analog AMPCPP at A-site | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, ... | | Authors: | Santos-Aledo, A, Plaza-Pegueroles, A, Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM uncovers a sequential mechanism for RNA polymerase I pausing and stalling at abasic DNA lesions.
Nat Commun, 16, 2025
|
|
9G2B
 
 | | Yeast RNA polymerase I elongation complex stalled by an apurinic site, 12-subunit | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Santos-Aledo, A, Plaza-Pegueroles, A, Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM uncovers a sequential mechanism for RNA polymerase I pausing and stalling at abasic DNA lesions.
Nat Commun, 16, 2025
|
|
9G24
 
 | | Yeast RNA polymerase I elongation complex stalled by an apurinic site bound to nucleotide analog AMPCPP at E-site | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, ... | | Authors: | Santos-Aledo, A, Plaza-Pegueroles, A, Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM uncovers a sequential mechanism for RNA polymerase I pausing and stalling at abasic DNA lesions.
Nat Commun, 16, 2025
|
|
9G29
 
 | | Yeast RNA polymerase I elongation complex stalled by an apurinic site with the C-terminal of A12 in the funnel | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Santos-Aledo, A, Plaza-Pegueroles, A, Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM uncovers a sequential mechanism for RNA polymerase I pausing and stalling at abasic DNA lesions.
Nat Commun, 16, 2025
|
|
9G2C
 
 | | Yeast RNA polymerase I elongation complex stalled by an apurinic site, open state | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA190, ... | | Authors: | Santos-Aledo, A, Plaza-Pegueroles, A, Ruiz, F.M, Fernandez-Tornero, C. | | Deposit date: | 2024-07-10 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM uncovers a sequential mechanism for RNA polymerase I pausing and stalling at abasic DNA lesions.
Nat Commun, 16, 2025
|
|
7AF8
 
 | | Bacterial 30S ribosomal subunit assembly complex state E (head domain) | | Descriptor: | 16SrRNA (head domain of the 30S ribosome, 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Fucini, P, Connell, S. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7AFR
 
 | | Ribosome maturation factor RimP (apo) | | Descriptor: | Ribosome maturation factor RimP | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Fucini, P, Connell, S. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7AFQ
 
 | | Ribosome binding factor A (RbfA) | | Descriptor: | Ribosome-binding factor A | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Fucini, P, Connell, S. | | Deposit date: | 2020-09-19 | | Release date: | 2020-12-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
4ZTD
 
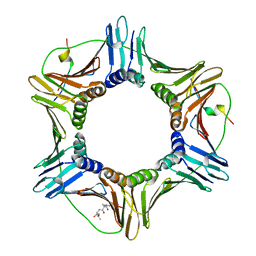 | | Crystal Structure of Human PCNA in complex with a TRAIP peptide | | Descriptor: | ALA-GLY-ALA-GLY-ALA, ALA-PHE-GLN-ALA-LYS-LEU-ASP-THR-PHE-LEU-TRP-SER, Proliferating cell nuclear antigen | | Authors: | Montoya, G, Mortuza, G.B, Blanco, F.J, Ibanez de Opakua, A. | | Deposit date: | 2015-05-14 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | TRAIP is a PCNA-binding ubiquitin ligase that protects genome stability after replication stress.
J.Cell Biol., 212, 2016
|
|
