5JCZ
 
 | | Rab11 bound to MyoVa-GTD | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Pylypenko, O, Attanda, W, Gauquelin, C, Malherbes, G, Houdusse, A. | | Deposit date: | 2016-04-15 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.056 Å) | | Cite: | Coordinated recruitment of Spir actin nucleators and myosin V motors to Rab11 vesicle membranes.
Elife, 5, 2016
|
|
4WHY
 
 | |
4WHT
 
 | |
2YK0
 
 | | Structure of the N-terminal NTS-DBL1-alpha and CIDR-gamma double domain of the PfEMP1 protein from Plasmodium falciparum varO strain. | | Descriptor: | ERYTHROCYTE MEMBRANE PROTEIN 1, MAGNESIUM ION, SULFATE ION | | Authors: | Lewit-Bentley, A, Juillerat, A, Vigan-Womas, I, Guillotte, M, Hessel, A, Raynal, B, Mercereau-Puijalon, O, Bentley, G.A. | | Deposit date: | 2011-05-25 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Abo Blood-Group Dependence of Plasmodium Falciparum Rosetting.
Plos Pathog., 8, 2012
|
|
5JCY
 
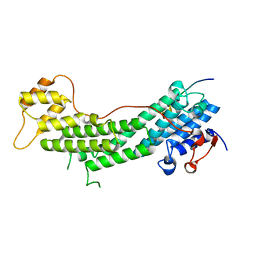 | | Spir2-GTBM bound to MyoVa-GTD | | Descriptor: | Protein spire homolog 2, Unconventional myosin-Va | | Authors: | Pylypenko, O, Malherbes, G, Welz, T, Kerkhoff, E, Houdusse, A. | | Deposit date: | 2016-04-15 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Coordinated recruitment of Spir actin nucleators and myosin V motors to Rab11 vesicle membranes.
Elife, 5, 2016
|
|
5LXL
 
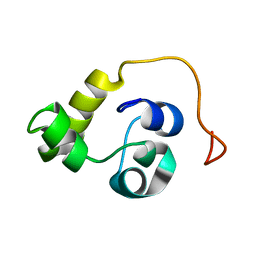 | | NMR structure of the N-terminal domain of the Bacteriophage T5 decoration protein pb10 | | Descriptor: | Decoration protein | | Authors: | Vernhes, E, Gilquin, B, Cuniasse, P, Boulanger, P, Zinn-Justin, S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid.
Sci Rep, 7, 2017
|
|
5LXK
 
 | | NMR structure of the C-terminal domain of the Bacteriophage T5 decoration protein pb10. | | Descriptor: | Decoration protein | | Authors: | Vernhes, E, Gilquin, B, Cuniasse, P, Boulanger, P, Zinn-Justin, S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid.
Sci Rep, 7, 2017
|
|
5LCV
 
 | |
7QRG
 
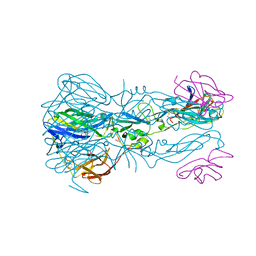 | |
7QRE
 
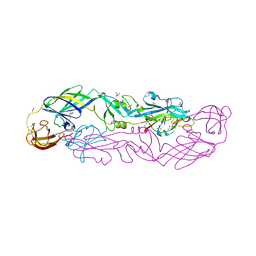 | | Structure of the hetero-tetramer complex between precursor membrane protein fragment (pr) and envelope protein (E) from tick-borne encephalitis virus | | Descriptor: | ACETATE ION, Envelope protein E, Genome polyprotein, ... | | Authors: | Vaney, M.C, Dellarole, M, Rey, F.A. | | Deposit date: | 2022-01-11 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Evolution and activation mechanism of the flavivirus class II membrane-fusion machinery.
Nat Commun, 13, 2022
|
|
7QRF
 
 | | Structure of the dimeric complex between precursor membrane ectodomain (prM) and envelope protein ectodomain (E) from tick-borne encephalitis virus | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Envelope protein E, ... | | Authors: | Vaney, M.C, Rouvinski, A, Rey, F.A. | | Deposit date: | 2022-01-11 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Evolution and activation mechanism of the flavivirus class II membrane-fusion machinery.
Nat Commun, 13, 2022
|
|
4LX2
 
 | | Crystal structure of Myo5a globular tail domain in complex with melanophilin GTBD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Melanophilin, Unconventional myosin-Va | | Authors: | Pylypenko, O, Attanda, W, Gauquelin, C, Houdusse, A. | | Deposit date: | 2013-07-29 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of myosin V Rab GTPase-dependent cargo recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LX0
 
 | | Crystal structure of Myo5b globular tail domain in complex with active Rab11a | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Pylypenko, O, Attanda, W, Gauquelin, C, Houdusse, A. | | Deposit date: | 2013-07-29 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis of myosin V Rab GTPase-dependent cargo recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LX1
 
 | | Crystal structure of Myo5a globular tail domain | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Pylypenko, O, Attanda, W, Coulibaly, D, Gauquelin, C, Houdusse, A. | | Deposit date: | 2013-07-29 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis of myosin V Rab GTPase-dependent cargo recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LWZ
 
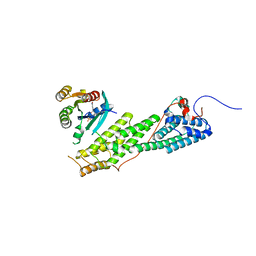 | | Crystal structure of Myo5b globular tail domain in complex with inactive Rab11a | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-11A, ... | | Authors: | Pylypenko, O, Attanda, W, Gauquelin, C, Houdusse, A. | | Deposit date: | 2013-07-29 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of myosin V Rab GTPase-dependent cargo recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5LK2
 
 | |
5LJZ
 
 | | Structure of hantavirus envelope glycoprotein Gc in postfusion conformation | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein, ... | | Authors: | Guardado-Calvo, P, Rey, F.A. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanistic Insight into Bunyavirus-Induced Membrane Fusion from Structure-Function Analyses of the Hantavirus Envelope Glycoprotein Gc.
Plos Pathog., 12, 2016
|
|
5LJY
 
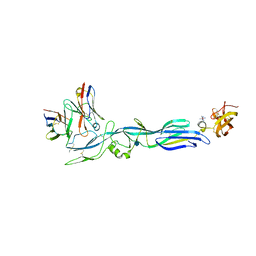 | | Structure of hantavirus envelope glycoprotein Gc in complex with scFv A5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT HEXAMMINE(III), Envelopment polyprotein, ... | | Authors: | Guardado-Calvo, P, Stettner, E, Jeffers, S.A, Rey, F.A. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanistic Insight into Bunyavirus-Induced Membrane Fusion from Structure-Function Analyses of the Hantavirus Envelope Glycoprotein Gc.
Plos Pathog., 12, 2016
|
|
8SWI
 
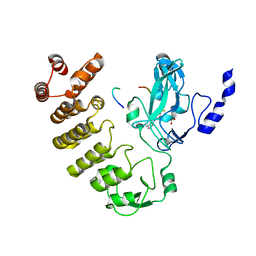 | |
8SR6
 
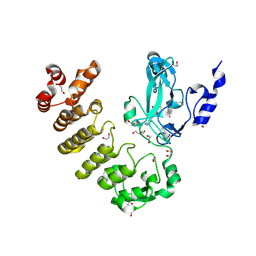 | | Crystal structure of legAS4 from Legionella pneumophila subsp. pneumophila with histone H3 (3-17)peptide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Eukaryotic huntingtin interacting protein B, ... | | Authors: | Xu, C, Chung, I.Y.W, Cygler, M. | | Deposit date: | 2023-05-05 | | Release date: | 2024-03-13 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The SET and ankyrin domains of the secreted Legionella pneumophila histone methyltransferase work together to modify host chromatin.
Mbio, 14, 2023
|
|
7BES
 
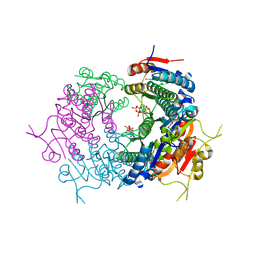 | | CryoEM structure of Mycobacterium tuberculosis UMP Kinase (UMPK) in complex with UDP and UTP | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, URIDINE-5'-DIPHOSPHATE, Uridylate kinase | | Authors: | Bous, J, Trapani, S, Walter, P, Bron, P, Munier-Lehmann, H. | | Deposit date: | 2020-12-24 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for the allosteric inhibition of UMP kinase from Gram-positive bacteria, a promising antibacterial target.
Febs J., 289, 2022
|
|
5LK3
 
 | |
7BL7
 
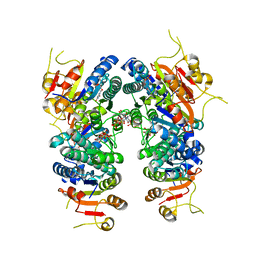 | | Crystal structure of UMPK from M. tuberculosis in complex with UDP and UTP (P21212 form) | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, URIDINE-5'-DIPHOSPHATE, Uridylate kinase | | Authors: | Walter, P, Labesse, G, Haouz, A, Mechaly, A.E, Munier-Lehmann, H. | | Deposit date: | 2021-01-18 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Structural basis for the allosteric inhibition of UMP kinase from Gram-positive bacteria, a promising antibacterial target.
Febs J., 289, 2022
|
|
7BIX
 
 | | Crystal structure of UMPK from M. tuberculosis in complex with UDP and UTP (C2 form) | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, URIDINE-5'-DIPHOSPHATE, Uridylate kinase | | Authors: | Labesse, G, Walter, P, Haouz, A, Mechaly, A.E, Munier-Lehmann, H. | | Deposit date: | 2021-01-13 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structural basis for the allosteric inhibition of UMP kinase from Gram-positive bacteria, a promising antibacterial target.
Febs J., 289, 2022
|
|
6T7Y
 
 | | Structure of PCNA bound to cPIP motif of DP2 from P. abyssi | | Descriptor: | DNA polymerase sliding clamp, cPIP motif from the DP2 large subunit of PolD | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
