4P0P
 
 | | Crystal structure of Human Mus81-Eme1 in complex with 5'-flap DNA, and Mg2+ | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, DNA GAATGTGTGTCTCAATC, ... | | Authors: | Gwon, G.H, Baek, K, Cho, Y. | | Deposit date: | 2014-02-22 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the structure-selective nuclease Mus81-Eme1 bound to flap DNA substrates.
Embo J., 33, 2014
|
|
5E9F
 
 | | Structural insights of isocitrate lyases from Magnaporthe oryzae | | Descriptor: | Isocitrate lyase, MAGNESIUM ION | | Authors: | Park, Y, Cho, Y, Lee, Y.-H, Lee, Y.-W, Rhee, S. | | Deposit date: | 2015-10-15 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and functional analysis of isocitrate lyases from Magnaporthe oryzae and Fusarium graminearum
J.Struct.Biol., 194, 2016
|
|
5E9H
 
 | | Structural insights of isocitrate lyases from Fusarium graminearum | | Descriptor: | Isocitrate lyase, MALONATE ION, MANGANESE (II) ION | | Authors: | Park, Y, Cho, Y, Lee, Y.-H, Lee, Y.-W, Rhee, S. | | Deposit date: | 2015-10-15 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and functional analysis of isocitrate lyases from Magnaporthe oryzae and Fusarium graminearum
J.Struct.Biol., 194, 2016
|
|
5E9G
 
 | | Structural insights of isocitrate lyases from Magnaporthe oryzae | | Descriptor: | GLYCEROL, GLYOXYLIC ACID, Isocitrate lyase, ... | | Authors: | Park, Y, Cho, Y, Lee, Y.-H, Lee, Y.-W, Rhee, S. | | Deposit date: | 2015-10-15 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and functional analysis of isocitrate lyases from Magnaporthe oryzae and Fusarium graminearum
J.Struct.Biol., 194, 2016
|
|
4YGQ
 
 | | Crystal structure of HAD phosphatase from Thermococcus onnurineus | | Descriptor: | Hydrolase, TERTIARY-BUTYL ALCOHOL | | Authors: | Ngo, T.D, Le, B.V, Subramani, V.K, Nguyen, C.M.T, Lee, H.S, Cho, Y, Kim, K.K, Hwang, H.Y. | | Deposit date: | 2015-02-26 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the substrate selectivity of a HAD phosphatase from Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 461, 2015
|
|
4YGR
 
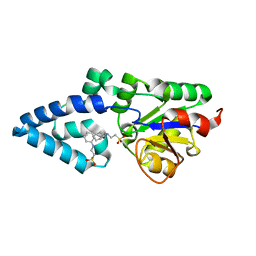 | | Crystal structure of HAD phosphatase from Thermococcus onnurineus | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hydrolase, MAGNESIUM ION | | Authors: | Ngo, T.D, Le, B.V, Subramani, V.K, Nguyen, C.M.T, Lee, H.S, Cho, Y, Kim, K.K, Hwang, H.Y. | | Deposit date: | 2015-02-26 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Structural basis for the substrate selectivity of a HAD phosphatase from Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 461, 2015
|
|
3T1I
 
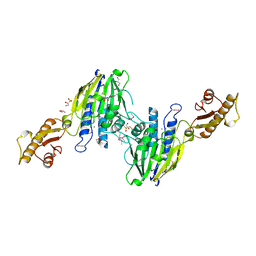 | | Crystal Structure of Human Mre11: Understanding Tumorigenic Mutations | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Double-strand break repair protein MRE11A, GLYCEROL, ... | | Authors: | Park, Y.B, Chae, J, Kim, Y, Cho, Y. | | Deposit date: | 2011-07-22 | | Release date: | 2011-11-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human mre11: understanding tumorigenic mutations
Structure, 19, 2011
|
|
4YGS
 
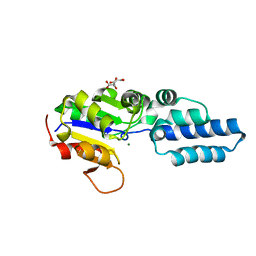 | | Crystal structure of HAD phosphatase from Thermococcus onnurineus | | Descriptor: | CITRIC ACID, Hydrolase, MAGNESIUM ION | | Authors: | Ngo, T.D, Le, B.V, Subramani, V.K, Nguyen, C.M.T, Lee, H.S, Cho, Y, Kim, K.K, Hwang, H.Y. | | Deposit date: | 2015-02-26 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the substrate selectivity of a HAD phosphatase from Thermococcus onnurineus NA1
Biochem.Biophys.Res.Commun., 461, 2015
|
|
5GOX
 
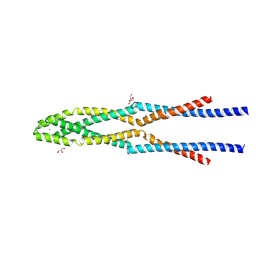 | | Eukaryotic Rad50 Functions as A Rod-shaped Dimer | | Descriptor: | DNA repair protein RAD50, GLYCEROL, ZINC ION | | Authors: | Park, Y.B, Hohl, M, Padjasek, M, Jeong, E, Jin, K.S, Krezel, A, Petrini, J.H.J, Cho, Y. | | Deposit date: | 2016-07-30 | | Release date: | 2017-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Eukaryotic Rad50 functions as a rod-shaped dimer
Nat. Struct. Mol. Biol., 24, 2017
|
|
1A77
 
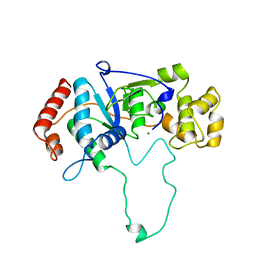 | | FLAP ENDONUCLEASE-1 FROM METHANOCOCCUS JANNASCHII | | Descriptor: | FLAP ENDONUCLEASE-1 PROTEIN, MAGNESIUM ION | | Authors: | Hwang, K.Y, Baek, K, Kim, H, Cho, Y. | | Deposit date: | 1998-03-20 | | Release date: | 1999-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of flap endonuclease-1 from Methanococcus jannaschii.
Nat.Struct.Biol., 5, 1998
|
|
1AD6
 
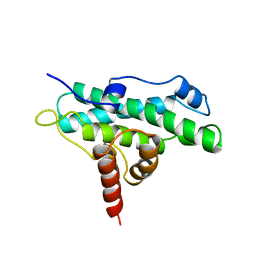 | | DOMAIN A OF HUMAN RETINOBLASTOMA TUMOR SUPPRESSOR | | Descriptor: | RETINOBLASTOMA TUMOR SUPPRESSOR | | Authors: | Kim, H.Y, Cho, Y. | | Deposit date: | 1997-02-21 | | Release date: | 1998-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural similarity between the pocket region of retinoblastoma tumour suppressor and the cyclin-box.
Nat.Struct.Biol., 4, 1997
|
|
1A76
 
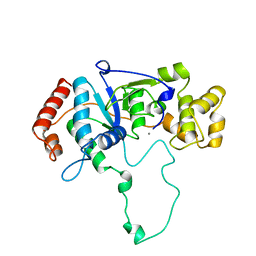 | | FLAP ENDONUCLEASE-1 FROM METHANOCOCCUS JANNASCHII | | Descriptor: | FLAP ENDONUCLEASE-1 PROTEIN, MANGANESE (II) ION | | Authors: | Hwang, K.Y, Baek, K, Kim, H, Cho, Y. | | Deposit date: | 1998-03-20 | | Release date: | 1999-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of flap endonuclease-1 from Methanococcus jannaschii.
Nat.Struct.Biol., 5, 1998
|
|
1B78
 
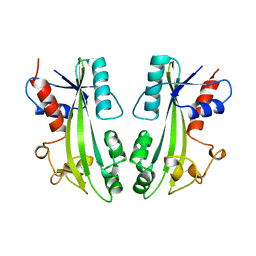 | | STRUCTURE-BASED IDENTIFICATION OF THE BIOCHEMICAL FUNCTION OF A HYPOTHETICAL PROTEIN FROM METHANOCOCCUS JANNASCHII:MJ0226 | | Descriptor: | PYROPHOSPHATASE | | Authors: | Hwang, K.Y, Chung, J.H, Han, Y.S, Kim, S.H, Cho, Y. | | Deposit date: | 1999-01-27 | | Release date: | 2000-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based identification of a novel NTPase from Methanococcus jannaschii.
Nat.Struct.Biol., 6, 1999
|
|
1B8V
 
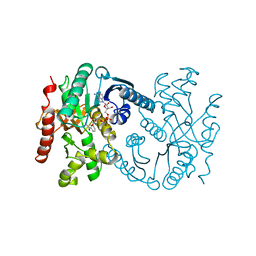 | | Malate dehydrogenase from Aquaspirillum arcticum | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (MALATE DEHYDROGENASE) | | Authors: | Kim, S.Y, Hwang, K.Y, Kim, S.H, Han, Y.S, Cho, Y. | | Deposit date: | 1999-02-02 | | Release date: | 1999-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum.
J.Biol.Chem., 274, 1999
|
|
1B8U
 
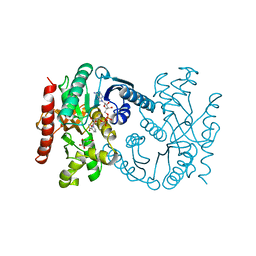 | | MALATE DEHYDROGENASE FROM AQUASPIRILLUM ARCTICUM | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXALOACETATE ION, PROTEIN (MALATE DEHYDROGENASE) | | Authors: | Kim, S.Y, Hwang, K.Y, Kim, S.-H, Han, Y.S, Cho, Y. | | Deposit date: | 1999-02-02 | | Release date: | 1999-07-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum.
J.Biol.Chem., 274, 1999
|
|
1B8P
 
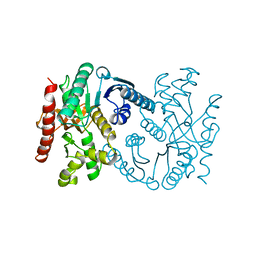 | | MALATE DEHYDROGENASE FROM AQUASPIRILLUM ARCTICUM | | Descriptor: | PROTEIN (MALATE DEHYDROGENASE) | | Authors: | Kim, S.Y, Hwang, K.Y, Kim, S.-H, Han, Y.S, Cho, Y. | | Deposit date: | 1999-02-02 | | Release date: | 1999-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum.
J.Biol.Chem., 274, 1999
|
|
5X06
 
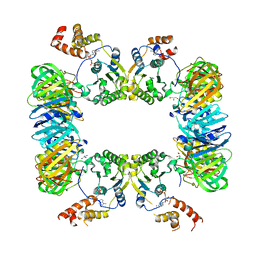 | | DNA replication regulation protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase III subunit beta, DnaA regulatory inactivator Hda, ... | | Authors: | Kim, J, Cho, Y. | | Deposit date: | 2017-01-20 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.237 Å) | | Cite: | Replication regulation protein
To Be Published
|
|
3TAL
 
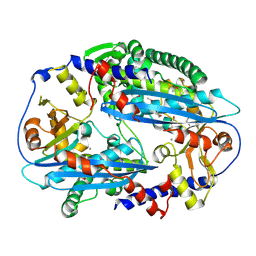 | | Crystal structure of NurA with manganese | | Descriptor: | DNA double-strand break repair protein nurA, GLYCEROL, MANGANESE (II) ION | | Authors: | Chae, J, Kim, Y.C, Cho, Y. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of the NurA-dAMP-Mn2+ complex
Nucleic Acids Res., 40, 2012
|
|
3TAZ
 
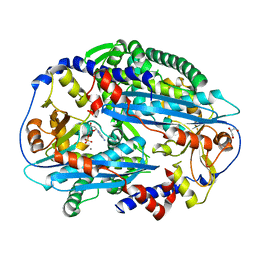 | | Crystal structure of NurA bound to dAMP and manganese | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, DNA double-strand break repair protein nurA, GLYCEROL, ... | | Authors: | Chae, J, Kim, Y.C, Cho, Y. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the NurA-dAMP-Mn2+ complex
Nucleic Acids Res., 40, 2012
|
|
3TAI
 
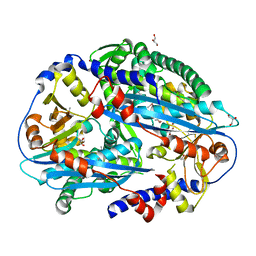 | | Crystal structure of NurA | | Descriptor: | DNA double-strand break repair protein nurA, GLYCEROL | | Authors: | Chae, J, Kim, Y.C, Cho, Y. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal structure of the NurA-dAMP-Mn2+ complex
Nucleic Acids Res., 40, 2012
|
|
5Y7Q
 
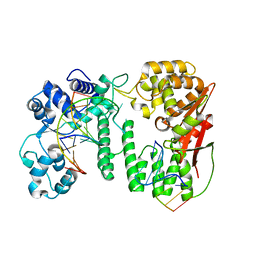 | | Crystal structure of paFAN1 bound to 2nt 5'flap DNA with gap | | Descriptor: | DNA (5'-D(*TP*TP*CP*AP*CP*AP*CP*AP*TP*TP*CP*AP*A)-3'), DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*AP*CP*TP*T)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Jin, H, Cho, Y. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural mechanism of DNA interstrand cross-link unhooking by the bacterial FAN1 nuclease.
J. Biol. Chem., 293, 2018
|
|
5Z6W
 
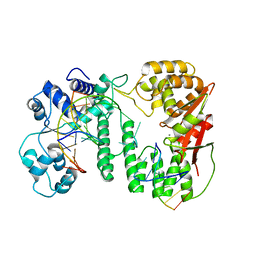 | | Crystal structure of paFAN1 bound to 2nt 5'flap DNA with gap with Manganese | | Descriptor: | DNA (5'-D(P*AP*TP*TP*CP*AP*A)-3'), DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*AP*CP*TP*T)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Jin, H, Cho, Y. | | Deposit date: | 2018-01-25 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural mechanism of DNA interstrand cross-link unhooking by the bacterial FAN1 nuclease.
J. Biol. Chem., 293, 2018
|
|
1COJ
 
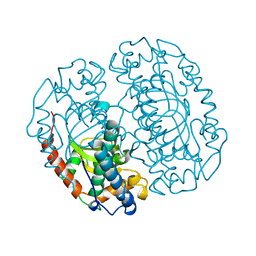 | | FE-SOD FROM AQUIFEX PYROPHILUS, A HYPERTHERMOPHILIC BACTERIUM | | Descriptor: | FE (III) ION, PROTEIN (SUPEROXIDE DISMUTASE) | | Authors: | Lim, J.H, Yu, Y.G, Kim, S.-H, Cho, S.-J, Ahn, B.Y, Han, Y.S, Cho, Y. | | Deposit date: | 1999-05-28 | | Release date: | 1999-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of an Fe-superoxide dismutase from the hyperthermophile Aquifex pyrophilus at 1.9 A resolution: structural basis for thermostability.
J.Mol.Biol., 270, 1997
|
|
9IMA
 
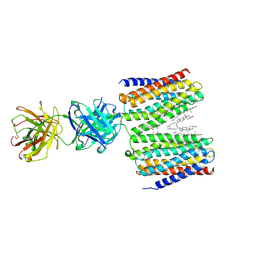 | | Cryo-EM structure for the GPRC5D complexed with Talquetamab Fab | | Descriptor: | CHOLESTEROL, G-protein coupled receptor family C group 5 member D, Talquetamab Fab (anti-GPRC5D) Heavy chain, ... | | Authors: | Jeong, J, Shin, J, Park, J, Cho, Y. | | Deposit date: | 2024-07-02 | | Release date: | 2024-09-11 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural Basis for the Recognition of GPRC5D by Talquetamab, a Bispecific Antibody for Multiple Myeloma.
J.Mol.Biol., 436, 2024
|
|
9JT7
 
 | | SFX reaction state structure (0-60min) of alanine racemase | | Descriptor: | ALANINE, Alanine racemase 2, CHLORIDE ION, ... | | Authors: | Kim, J, Nam, K.H, Cho, Y. | | Deposit date: | 2024-10-02 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploring the reaction dynamics of alanine racemase using serial femtosecond crystallography.
Sci Rep, 14, 2024
|
|
