5BQF
 
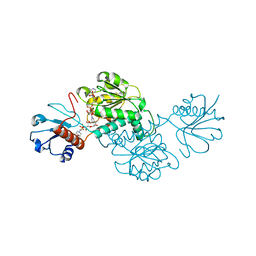 | | Probable 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADP, HEPES and L(+)-tartaric acid | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Langner, K.M, Shabalin, I.G, Handing, K.B, Gasiorowska, O.A, Stead, M, Hillerich, B.S, Chowdhury, S, Hammonds, J, Zimmerman, M.D, Al Obaidi, N, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-05-29 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Probable 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADP, HEPES and L-tartaric acid
to be published
|
|
4WBT
 
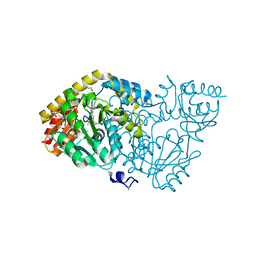 | | Crystal structure of histidinol-phosphate aminotransferase from Sinorhizobium meliloti in complex with pyridoxal-5'-phosphate | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Shabalin, I.G, Bacal, P, Kowalska, A.K, Cooper, D.R, Stead, M, Hammonds, J, Ahmed, M, Hillerich, B.S, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-03 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of histidinol-phosphate aminotransferase from Sinorhizobium meliloti in complex with pyridoxal-5'-phosphate
to be published
|
|
4XCV
 
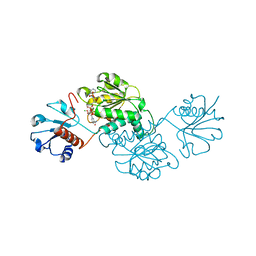 | | Probable 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADPH | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, NADP-dependent 2-hydroxyacid dehydrogenase, ... | | Authors: | Langner, K.M, Shabalin, I.G, Handing, K.B, Gasiorowska, O.A, Stead, M, Hillerich, B.S, Chowdhury, S, Hammonds, J, Zimmerman, M.D, Al Obadi, N, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-12-18 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADPH
to be published
|
|
5EPE
 
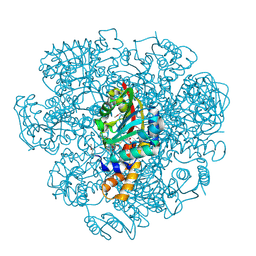 | | Crystal structure of SAM-dependent methyltransferase from Thiobacillus denitrificans in complex with S-Adenosyl-L-homocysteine | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase, SODIUM ION | | Authors: | LaRowe, C, Shabalin, I.G, Kutner, J, Handing, K.B, Stead, M, Hillerich, B.S, Ahmed, M, Seidel, R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-11-25 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase from Thiobacillus denitrificans in complex with S-Adenosyl-L-homocysteine
to be published
|
|
4HKT
 
 | |
4HC8
 
 | |
3MKV
 
 | | Crystal structure of amidohydrolase eaj56179 | | Descriptor: | CARBONATE ION, GLYCEROL, PUTATIVE AMIDOHYDROLASE, ... | | Authors: | Patskovsky, Y, Bonanno, J, Ozyurt, S, Sauder, J.M, Freeman, J, Wu, B, Smith, D, Bain, K, Rodgers, L, Wasserman, S.R, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-15 | | Release date: | 2010-04-28 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
4EJM
 
 | |
4EJ6
 
 | |
4E6P
 
 | |
4G8S
 
 | | Crystal Structure Of a Putative Nitroreductase from Geobacter sulfurreducens PCA (Target PSI-013445) | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Nitroreductase family protein | | Authors: | Kumar, P.R, Bhosle, R, Hillerich, B, Seidel, R, Toro, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a nitroreductase from Geobacter sulfurreducens PCA
to be published
|
|
4HPS
 
 | |
4HKM
 
 | |
4XK2
 
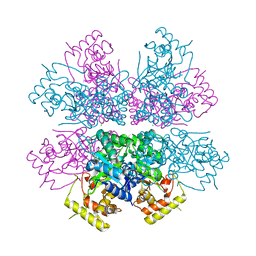 | | Crystal structure of aldo-keto reductase from Polaromonas sp. JS666 | | Descriptor: | Aldo/keto reductase, CHLORIDE ION, SODIUM ION | | Authors: | Gasiorowska, O.A, Handing, K.B, Shabalin, I.G, Sroka, P, Hillerich, B.S, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-01-09 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of aldo-keto reductase from Polaromonas sp. JS666
to be published
|
|
4JWT
 
 | |
3HPF
 
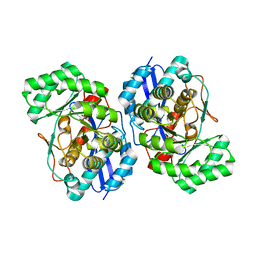 | | Crystal structure of the mutant Y90F of divergent galactarate dehydratase from Oceanobacillus iheyensis complexed with Mg and galactarate | | Descriptor: | D-galactaric acid, MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-06-04 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
4LDN
 
 | |
3FYY
 
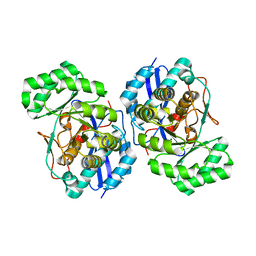 | | Crystal structure of divergent enolase from Oceanobacillus iheyensis complexed with Mg | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
4L0M
 
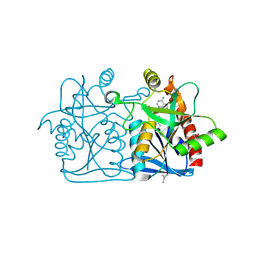 | |
4KN5
 
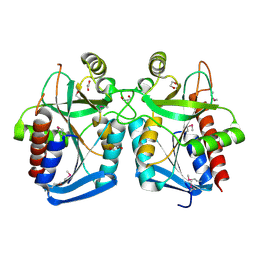 | |
4JOS
 
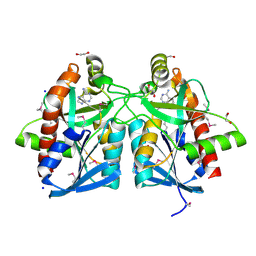 | | Crystal structure of a putative 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Francisella philomiragia ATCC 25017 (Target NYSGRC-029335) | | Descriptor: | 1,2-ETHANEDIOL, ADENINE, Adenosylhomocysteine nucleosidase, ... | | Authors: | Sampathkumar, P, Schramm, V.L, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-18 | | Release date: | 2013-04-03 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a putative 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Francisella philomiragia ATCC 25017 (Target NYSGRC-029335)
to be published
|
|
5V7N
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with NADP and 2-Keto-D-gluconic acid | | Descriptor: | 2-keto-D-gluconic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shabalin, I.G, Handing, K.B, Miks, C.D, Kutner, J, Matelska, D, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-03-20 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5V7G
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with NADPH and oxalate | | Descriptor: | CHLORIDE ION, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Shabalin, I.G, Mason, D.V, Handing, K.B, Kutner, J, Matelska, D, Cooper, D.R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-03-20 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5UNN
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc02828 (SmGhrA) from Sinorhizobium meliloti in apo form | | Descriptor: | CHLORIDE ION, GLYCEROL, NADPH-dependent glyoxylate/hydroxypyruvate reductase | | Authors: | Shabalin, I.G, LaRowe, C, Kutner, J, Gasiorowska, O.A, Handing, K.B, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-01-31 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
4ESK
 
 | |
