5OE5
 
 | |
4PIM
 
 | |
4PIN
 
 | | Ergothioneine-biosynthetic methyltransferase EgtD in complex with N,N-dimethylhistidine | | Descriptor: | Histidine-specific methyltransferase EgtD, N,N-dimethyl-L-histidine, PHOSPHATE ION | | Authors: | Vit, A, Seebeck, F.P, Blankenfeldt, W. | | Deposit date: | 2014-05-09 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ergothioneine Biosynthetic Methyltransferase EgtD Reveals the Structural Basis of Aromatic Amino Acid Betaine Biosynthesis.
Chembiochem, 16, 2015
|
|
3B4O
 
 | | Crystal structure of phenazine biosynthesis protein PhzA/B from Burkholderia cepacia R18194, apo form | | Descriptor: | ACETATE ION, Phenazine biosynthesis protein A/B | | Authors: | Ahuja, E.G, Janning, P, Mentel, M, Graebsch, A, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2007-10-24 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
4PIO
 
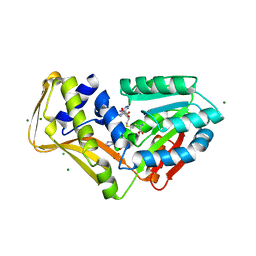 | | Ergothioneine-biosynthetic methyltransferase EgtD in complex with N,N-dimethylhistidine and SAH | | Descriptor: | CHLORIDE ION, Histidine-specific methyltransferase EgtD, MAGNESIUM ION, ... | | Authors: | Vit, A, Seebeck, F.P, Blankenfeldt, W. | | Deposit date: | 2014-05-09 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.506 Å) | | Cite: | Ergothioneine Biosynthetic Methyltransferase EgtD Reveals the Structural Basis of Aromatic Amino Acid Betaine Biosynthesis.
Chembiochem, 16, 2015
|
|
3B4P
 
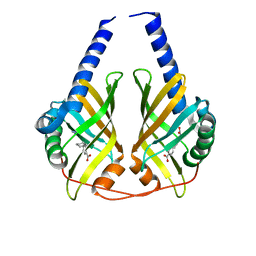 | | Crystal structure of phenazine biosynthesis protein PhzA/B from Burkholderia cepacia R18194, complex with 2-(cyclohexylamino)benzoic acid | | Descriptor: | 2-(cyclohexylamino)benzoic acid, ACETATE ION, AZIDE ION, ... | | Authors: | Ahuja, E.G, Janning, P, Mentel, M, Graebsch, A, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2007-10-24 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
4PIP
 
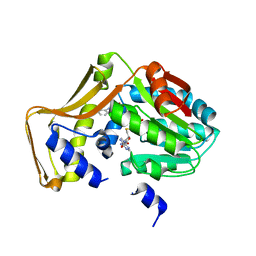 | | Engineered EgtD variant EgtD-M252V,E282A in complex with tryptophan and SAH | | Descriptor: | CHLORIDE ION, Histidine-specific methyltransferase EgtD, MAGNESIUM ION, ... | | Authors: | Vit, A, Seebeck, F.P, Blankenfeldt, W. | | Deposit date: | 2014-05-09 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ergothioneine Biosynthetic Methyltransferase EgtD Reveals the Structural Basis of Aromatic Amino Acid Betaine Biosynthesis.
Chembiochem, 16, 2015
|
|
6TUM
 
 | |
1KD0
 
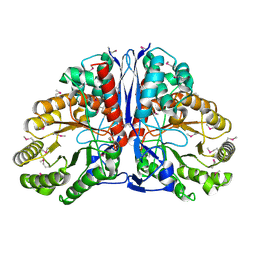 | | Crystal Structure of beta-methylaspartase from Clostridium tetanomorphum. Apo-structure. | | Descriptor: | 1,2-ETHANEDIOL, beta-methylaspartase | | Authors: | Asuncion, M, Blankenfeldt, W, Barlow, J.N, Gani, D, Naismith, J.H. | | Deposit date: | 2001-11-12 | | Release date: | 2001-12-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of 3-methylaspartase from Clostridium tetanomorphum functions via the common enolase chemical step.
J.Biol.Chem., 277, 2002
|
|
1KCZ
 
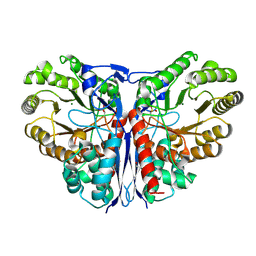 | | Crystal Structure of beta-methylaspartase from Clostridium tetanomorphum. Mg-complex. | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, beta-methylaspartase | | Authors: | Asuncion, M, Blankenfeldt, W, Barlow, J.N, Gani, D, Naismith, J.H. | | Deposit date: | 2001-11-12 | | Release date: | 2001-12-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of 3-methylaspartase from Clostridium tetanomorphum functions via the common enolase chemical step.
J.Biol.Chem., 277, 2002
|
|
6RTD
 
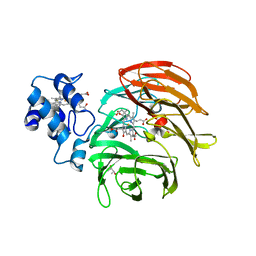 | | Dihydro-heme d1 dehydrogenase NirN in complex with DHE | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cytochrome c, HEME C, ... | | Authors: | Kluenemann, T, Preuss, A, Layer, G, Blankenfeldt, W. | | Deposit date: | 2019-05-23 | | Release date: | 2019-06-19 | | Last modified: | 2019-08-28 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal Structure of Dihydro-Heme d1Dehydrogenase NirN from Pseudomonas aeruginosa Reveals Amino Acid Residues Essential for Catalysis.
J.Mol.Biol., 431, 2019
|
|
6RTE
 
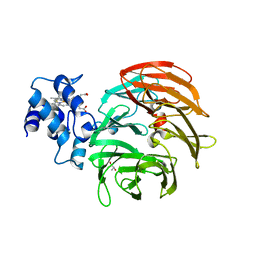 | | Dihydro-heme d1 dehydrogenase NirN in complex with DHE | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cytochrome c, HEME C | | Authors: | Kluenemann, T, Preuss, A, Layer, G, Blankenfeldt, W. | | Deposit date: | 2019-05-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Dihydro-Heme d1Dehydrogenase NirN from Pseudomonas aeruginosa Reveals Amino Acid Residues Essential for Catalysis.
J.Mol.Biol., 431, 2019
|
|
5MZW
 
 | | Crystal structure of the decarboxylase AibA/AibB | | Descriptor: | GLYCEROL, Glutaconate CoA-transferase family, subunit A, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MZY
 
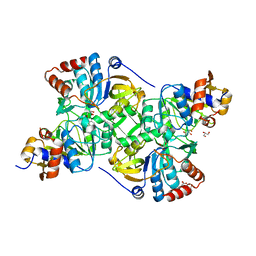 | | Crystal structure of the decarboxylase AibA/AibB in complex with a possible transition state analog | | Descriptor: | (1~{R},2~{S})-2-methylcyclohexane-1-carboxylic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5N00
 
 | | Crystal structure of the decarboxylase AibA/AibB C56A variant | | Descriptor: | ACETATE ION, Glutaconate CoA-transferase family, subunit A, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5NCH
 
 | | GriE apo form | | Descriptor: | Leucine hydroxylase | | Authors: | Lukat, P, Blankenfeldt, W, Mueller, R. | | Deposit date: | 2017-03-05 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Biosynthesis of methyl-proline containing griselimycins, natural products with anti-tuberculosis activity.
Chem Sci, 8, 2017
|
|
5N02
 
 | | Crystal structure of the decarboxylase AibA/AibB C56S variant | | Descriptor: | ACETATE ION, Glutaconate CoA-transferase family, subunit A, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5N03
 
 | | Crystal structure of the decarboxylase AibA/AibB C56V variant | | Descriptor: | ACETATE ION, Glutaconate CoA-transferase family, subunit A, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MZZ
 
 | | Crystal structure of the decarboxylase AibA/AibB in complex with 3-methylglutaconate | | Descriptor: | 3-methylpent-2-enedioic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
3JUQ
 
 | |
5NCI
 
 | | GriE in complex with cobalt, alpha-ketoglutarate and l-leucine | | Descriptor: | 2-OXOGLUTARIC ACID, COBALT (II) ION, LEUCINE, ... | | Authors: | Lukat, P, Blankenfeldt, W, Mueller, R. | | Deposit date: | 2017-03-05 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Biosynthesis of methyl-proline containing griselimycins, natural products with anti-tuberculosis activity.
Chem Sci, 8, 2017
|
|
3JUN
 
 | |
5N01
 
 | | Crystal structure of the decarboxylase AibA/AibB C56N variant | | Descriptor: | ACETATE ION, GLYCEROL, Glutaconate CoA-transferase family, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5NCJ
 
 | | GriE in complex with manganese, succinate and (2S,4R)-5-hydroxyleucine | | Descriptor: | (2S,4R)-5-hydroxyleucine, Leucine hydroxylase, MANGANESE (II) ION, ... | | Authors: | Lukat, P, Blankenfeldt, W, Mueller, R. | | Deposit date: | 2017-03-05 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.529 Å) | | Cite: | Biosynthesis of methyl-proline containing griselimycins, natural products with anti-tuberculosis activity.
Chem Sci, 8, 2017
|
|
3JUM
 
 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with 5-bromo-2-((1S,3R)-3-carboxycyclohexylamino)benzoic acid | | Descriptor: | 5-bromo-2-{[(1S,3R)-3-carboxycyclohexyl]amino}benzoic acid, Phenazine biosynthesis protein A/B | | Authors: | Mentel, M, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2009-09-15 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Active Site of an Enzyme Can Host Both Enantiomers of a Racemic Ligand Simultaneously
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
