5Y6A
 
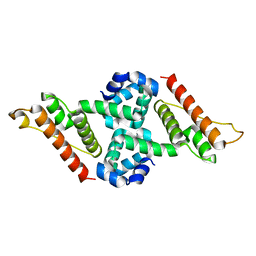 | | Crystal structure of the anti-CRISPR protein, AcrIIA1 | | Descriptor: | chain A and B | | Authors: | Ka, D, An, S.Y, Suh, J.Y, Bae, E. | | Deposit date: | 2017-08-11 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an anti-CRISPR protein, AcrIIA1
Nucleic Acids Res., 46, 2018
|
|
8PPK
 
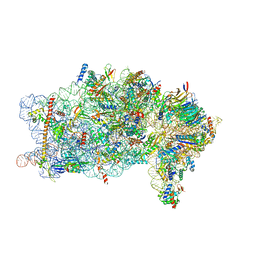 | | Bat-Hp-CoV Nsp1 and eIF1 bound to the human 40S small ribosomal subunit | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
8PPL
 
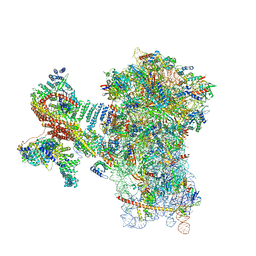 | | MERS-CoV Nsp1 bound to the human 43S pre-initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
2Q4W
 
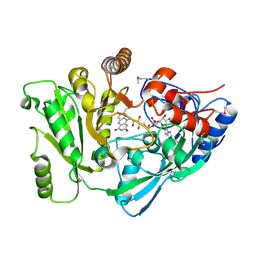 | | Ensemble refinement of the protein crystal structure of cytokinin oxidase/dehydrogenase (CKX) from Arabidopsis thaliana At5g21482 | | Descriptor: | Cytokinin dehydrogenase 7, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Arabidopsis thaliana cytokinin dehydrogenase.
Proteins, 70, 2008
|
|
2I2O
 
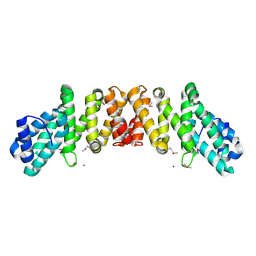 | | Crystal Structure of an eIF4G-like Protein from Danio rerio | | Descriptor: | NICKEL (II) ION, eIF4G-like protein | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-08-16 | | Release date: | 2006-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of an eIF4G-like protein from Danio rerio.
Proteins, 78, 2010
|
|
2Q53
 
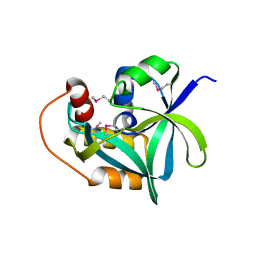 | | Ensemble refinement of the crystal structure of uncharacterized protein loc79017 from Homo sapiens | | Descriptor: | Uncharacterized protein C7orf24 | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of Homo sapiens protein LOC79017.
Proteins, 70, 2008
|
|
2GO9
 
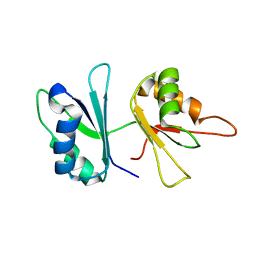 | | RRM domains 1 and 2 of Prp24 from S. cerevisiae | | Descriptor: | U4/U6 snRNA-associated splicing factor PRP24 | | Authors: | Reiter, N.J, Lee, D.H, Tonelli, M, Kwan, S.K, Brow, D.A, Butcher, S.E. | | Deposit date: | 2006-04-12 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions of the first three RNA recognition motifs of splicing factor prp24.
J.Mol.Biol., 367, 2007
|
|
1KI9
 
 | |
5XN4
 
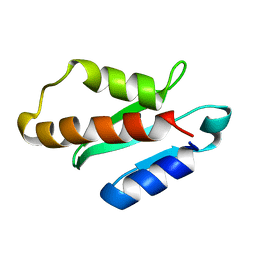 | | Anti-CRISPR protein AcrIIA4 | | Descriptor: | Anti-CRISPR AcrIIA4 | | Authors: | Suh, J.-Y, Kim, I. | | Deposit date: | 2017-05-17 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of anti-CRISPR AcrIIA4, the Cas9 inhibitor.
Sci Rep, 8, 2018
|
|
5CSR
 
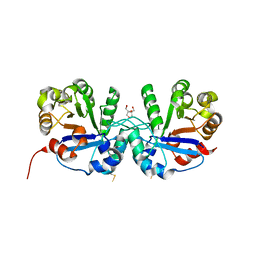 | | Crystal structure of triosephosphate isomerase from Thermoplasma acidophilium | | Descriptor: | CHLORIDE ION, GLYCEROL, Triosephosphate isomerase | | Authors: | Park, S.H, Kim, H.S, Song, M.K, Park, H.S, Han, B.W. | | Deposit date: | 2015-07-23 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure and Stability of the Dimeric Triosephosphate Isomerase from the Thermophilic Archaeon Thermoplasma acidophilum.
Plos One, 10, 2015
|
|
5CSS
 
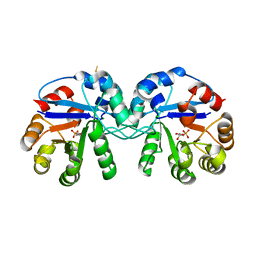 | | Crystal structure of triosephosphate isomerase from Thermoplasma acidophilum with glycerol 3-phosphate | | Descriptor: | CHLORIDE ION, SN-GLYCEROL-3-PHOSPHATE, Triosephosphate isomerase | | Authors: | Park, S.H, Kim, H.S, Song, M.K, Kim, K.R, Park, J.S, Han, B.W. | | Deposit date: | 2015-07-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and Stability of the Dimeric Triosephosphate Isomerase from the Thermophilic Archaeon Thermoplasma acidophilum.
Plos One, 10, 2015
|
|
3DL0
 
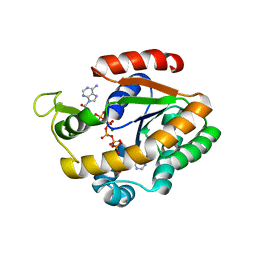 | | Crystal structure of adenylate kinase variant AKlse3 | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bannen, R.M, Bianchetti, C.M, Bingman, C.A, McCoy, J.G. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Effectiveness and limitations of local structural entropy optimization in the thermal stabilization of mesophilic and thermophilic adenylate kinases.
Proteins, 82, 2014
|
|
3DKV
 
 | | Crystal structure of adenylate kinase variant AKlse1 | | Descriptor: | 1,2-ETHANEDIOL, Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ... | | Authors: | Bannen, R.M, Bianchetti, C.M, Bingman, C.A, Bitto, E.B. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of adenylate kinase variant AKlse1.
To be Published
|
|
2ECK
 
 | | STRUCTURE OF PHOSPHOTRANSFERASE | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENYLATE KINASE | | Authors: | Berry, M.B, Bilderback, T, Glaser, M, Phillips Jr, G.N. | | Deposit date: | 1996-12-16 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ADP/AMP complex of Escherichia coli adenylate kinase.
Proteins, 62, 2006
|
|
6B6A
 
 | | Beta-Lactamase, 2secs timepoint, mixed, shards crystal form | | Descriptor: | (2R)-2-[(1S)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-hydroxyethyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, Ceftriaxone, ... | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
6B6C
 
 | |
6B6B
 
 | |
6B6E
 
 | | Beta-Lactamase, mixed with Ceftriaxone, needles crystal form, 500ms | | Descriptor: | (2R)-2-[(1S)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-hydroxyethyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, Ceftriaxone | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
6B68
 
 | | Beta-Lactamase, 100ms timepoint, mixed, shards crystal form | | Descriptor: | (2R)-2-[(1S)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-hydroxyethyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, Ceftriaxone, ... | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
1KHT
 
 | |
6B5X
 
 | | Beta-Lactamase, unmixed shards crystal form | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Pandey, S. | | Deposit date: | 2017-09-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
6B6F
 
 | |
6B69
 
 | | Beta-Lactamase, 500ms timepoint, mixed, shards crystal form | | Descriptor: | (2R)-2-[(1S)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-hydroxyethyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, (2R)-2-[(S)-{[(2E)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}(carboxy)methyl]-5-(hydroxymethyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, ... | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
6B6D
 
 | | Beta-Lactamase, mixed with Ceftriaxone, needles crystal form, 100ms | | Descriptor: | (2R)-2-[(1S)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-hydroxyethyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, Ceftriaxone | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
6B5Y
 
 | | Beta-lactamase, mixed with Ceftriaxone, 30ms time point, Shards crystal form | | Descriptor: | Beta-lactamase, Ceftriaxone, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-09-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
