6OKF
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and C16-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 1, Acyl carrier protein, N-[2-(dodecanoylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Mindrebo, J.T, Kim, W.E, Bartholow, T.G, Chen, A, Davis, T.D, La Clair, J, Burkart, M.D, Noel, J.P. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Gating mechanism of elongating beta-ketoacyl-ACP synthases.
Nat Commun, 11, 2020
|
|
6OLT
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabF, and C12-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, Acyl carrier protein, N-[2-(dodecanoylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide | | Authors: | Mindrebo, J.T, Kim, W.E, Bartholow, T.G, Chen, A, Davis, T.D, La Clair, J, Burkart, M.D, Noel, J.P. | | Deposit date: | 2019-04-17 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Gating mechanism of elongating beta-ketoacyl-ACP synthases.
Nat Commun, 11, 2020
|
|
4JYP
 
 | | crystal Structure of KAI2 Apo form | | Descriptor: | Hydrolase, alpha/beta fold family protein | | Authors: | Guo, Y, Zheng, Z, Noel, J.P. | | Deposit date: | 2013-03-31 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Smoke-derived karrikin perception by the alpha/beta-hydrolase KAI2 from Arabidopsis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JYM
 
 | | crystal Structure of KAI2 in complex with 3-methyl-2H-furo[2,3-c]pyran-2-one | | Descriptor: | 3-methyl-2H-furo[2,3-c]pyran-2-one, Hydrolase, alpha/beta fold family protein | | Authors: | Guo, Y, Zheng, Z, Noel, J.P. | | Deposit date: | 2013-03-30 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Smoke-derived karrikin perception by the alpha/beta-hydrolase KAI2 from Arabidopsis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6VKZ
 
 | |
6VL1
 
 | | Crystal Structure of the N-prenyltransferase DabA in Complex with NGG and Mg2+ | | Descriptor: | DabA, MAGNESIUM ION, N-[(2E)-3,7-dimethylocta-2,6-dien-1-yl]-L-glutamic acid | | Authors: | Chekan, J.R, Noel, J.P, Moore, B.S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Algal neurotoxin biosynthesis repurposes the terpene cyclase structural fold into anN-prenyltransferase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VL0
 
 | | Crystal Structure of the N-prenyltransferase DabA in Complex with GSPP and Mn2+ | | Descriptor: | DabA, GERANYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chekan, J.R, Noel, J.P, Moore, B.S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Algal neurotoxin biosynthesis repurposes the terpene cyclase structural fold into anN-prenyltransferase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5JNN
 
 | | Crystal structure of abscisic acid receptor PYL2 in complex with Phaseic acid. | | Descriptor: | (3S,4E)-5-[(1R,5R,8S)-8-hydroxy-1,5-dimethyl-3-oxo-6-oxabicyclo[3.2.1]octan-8-yl]-3-methylpent-4-enoic acid, Abscisic acid receptor PYL2 | | Authors: | Weng, J.K, Noel, J.P. | | Deposit date: | 2016-04-30 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Co-evolution of Hormone Metabolism and Signaling Networks Expands Plant Adaptive Plasticity.
Cell, 166, 2016
|
|
5JO7
 
 | | Henbane premnaspirodiene synthase (HPS), also known as Henbane vetispiradiene synthase (HVS) from Hyoscyamus muticus | | Descriptor: | Vetispiradiene synthase 1 | | Authors: | Koo, H.J, Xu, Y, Louie, G.V, Bowman, M, Noel, J.P. | | Deposit date: | 2016-05-02 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional study of terpene synthase from 512 mutant library of henbane premnaspirodiene synthase reveals protein residue interactions
To Be Published
|
|
5KJS
 
 | | Crystal Structure of Arabidopsis thaliana HCT | | Descriptor: | Shikimate O-hydroxycinnamoyltransferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KJV
 
 | | Crystal structure of Coleus blumei HCT | | Descriptor: | Hydroxycinnamoyl transferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KJT
 
 | | Crystal structure of Arabidopsis thaliana HCT in complex with p-coumaroyl-CoA | | Descriptor: | Shikimate O-hydroxycinnamoyltransferase, p-coumaroyl-CoA | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KJW
 
 | | Crystal structure of Coleus blumei HCT in complex with 3-hydroxyacetophenone | | Descriptor: | 1-(3-hydroxyphenyl)ethanone, Hydroxycinnamoyl transferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
5KJU
 
 | | Crystal structure of Arabidopsis thaliana HCT in complex with p-coumaroylshikimate | | Descriptor: | (3~{R},4~{S},5~{R})-3-[(~{E})-3-(4-hydroxyphenyl)prop-2-enoyl]oxy-4,5-bis(oxidanyl)cyclohexene-1-carboxylic acid, Shikimate O-hydroxycinnamoyltransferase | | Authors: | Levsh, O, Chiang, Y.C, Tung, C.F, Noel, J.P, Wang, Y, Weng, J.K. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Dynamic Conformational States Dictate Selectivity toward the Native Substrate in a Substrate-Permissive Acyltransferase.
Biochemistry, 55, 2016
|
|
6CIG
 
 | | CRYSTAL STRUCTURE ANALYSIS OF SELENOMETHIONINE SUBSTITUTED ISOFLAVONE O-METHYLTRANSFERASE | | Descriptor: | GLYCEROL, Isoflavone-7-O-methyltransferase 8, N-(TRIS(HYDROXYMETHYL)METHYL)-3-AMINOPROPANESULFONIC ACID, ... | | Authors: | Zubieta, C, Dixon, R.A, Shabalin, I.G, Kowiel, M, Porebski, P.J, Noel, J.P. | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-07 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases.
Nat. Struct. Biol., 8, 2001
|
|
6CJN
 
 | | Crystal Structure of Chalcone Isomerase from Medicago Sativa with the G95T mutation | | Descriptor: | Chalcone--flavonone isomerase 1, SULFATE ION | | Authors: | Burke, J.R, La Clair, J.J, Philippe, R.N, Pabis, A, Jez, J.M, Cortina, G, Kaltenbach, M, Bowman, M.E, Woods, K.B, Nelson, A.T, Tawfik, D.S, Kamerlin, S.C.L, Noel, J.P. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bifunctional Substrate Activation via an Arginine Residue Drives Catalysis in Chalcone Isomerases
Acs Catalysis, 2019
|
|
6CJO
 
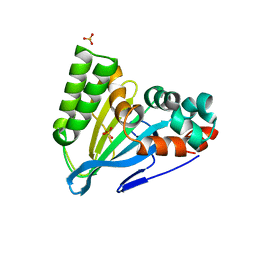 | | Crystal Structure of Chalcone Isomerase from Medicago Sativa with the G95S mutation. | | Descriptor: | Chalcone--flavonone isomerase 1, SULFATE ION | | Authors: | Burke, J.R, La Clair, J.J, Philippe, R.N, Pabis, A, Jez, J.M, Cortina, G, Kaltenbach, M, Bowman, M.E, Woods, K.B, Nelson, A.T, Tawfik, D.S, Kamerlin, S.C.L, Noel, J.P. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bifunctional Substrate Activation via an Arginine Residue Drives Catalysis in Chalcone Isomerases
Acs Catalysis, 2019
|
|
2F82
 
 | |
2FA0
 
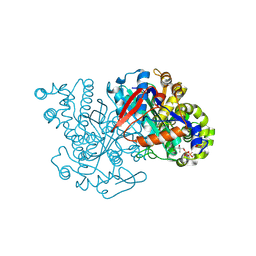 | | HMG-CoA synthase from Brassica juncea in complex with HMG-CoA and covalently bound to HMG-CoA | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, HMG-CoA synthase | | Authors: | Pojer, F, Ferrer, J.L, Richard, S.B, Noel, J.P. | | Deposit date: | 2005-12-06 | | Release date: | 2006-07-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis for the design of potent and species-specific inhibitors of 3-hydroxy-3-methylglutaryl CoA synthases.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2F9A
 
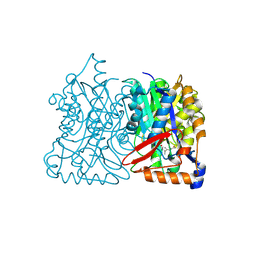 | | HMG-CoA synthase from Brassica juncea in complex with F-244 | | Descriptor: | (7R,12R,13R)-13-formyl-12,14-dihydroxy-3,5,7-trimethyltetradeca-2,4-dienoic acid, 3-Hydroxy-3-methylglutaryl coenzyme A synthase 1 | | Authors: | Pojer, F, Ferrer, J.L, Richard, S.B, Noel, J.P. | | Deposit date: | 2005-12-05 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis for the design of potent and species-specific inhibitors of 3-hydroxy-3-methylglutaryl CoA synthases.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2F21
 
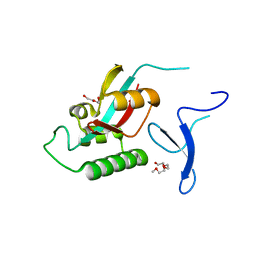 | | human Pin1 Fip mutant | | Descriptor: | PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jager, M, Zhang, Y, Bowman, M.E, Noel, J.P, Kelly, J.W. | | Deposit date: | 2005-11-15 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-function-folding relationship in a WW domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FA3
 
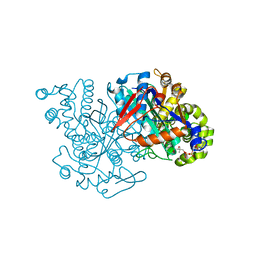 | | HMG-CoA synthase from Brassica juncea in complex with acetyl-CoA and acetyl-cys117. | | Descriptor: | ACETYL COENZYME *A, HMG-CoA synthase | | Authors: | Pojer, F, Ferrer, J.L, Richard, S.B, Noel, J.P. | | Deposit date: | 2005-12-06 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural basis for the design of potent and species-specific inhibitors of 3-hydroxy-3-methylglutaryl CoA synthases.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3W36
 
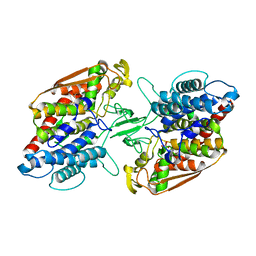 | | Crystal structure of holo-type bacterial Vanadium-dependent chloroperoxidase | | Descriptor: | NapH1, VANADATE ION | | Authors: | Liscombe, D.K, Miyanaga, A, Fielding, E, Bernhardt, P, Li, A, Winter, J.M, Gilson, M.K, Noel, J.P, Moore, B.S. | | Deposit date: | 2012-12-11 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis of Stereospecific Vanadium-Dependent Haloperoxidase Family Enzymes in Napyradiomycin Biosynthesis.
Biochemistry, 2022
|
|
3W8W
 
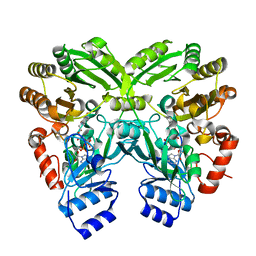 | | The crystal structure of EncM | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-dependent oxygenase EncM | | Authors: | Teufel, R, Miyanaga, A, Stull, F, Michaudel, Q, Louie, G, Noel, J.P, Baran, P.S, Palfey, B, Moore, B.S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Flavin-mediated dual oxidation controls an enzymatic Favorskii-type rearrangement.
Nature, 503, 2013
|
|
3W35
 
 | | Crystal structure of apo-type bacterial Vanadium-dependent chloroperoxidase | | Descriptor: | NapH1 | | Authors: | Liscombe, D.K, Miyanaga, A, Fielding, E, Bernhardt, P, Li, A, Winter, J.M, Gilson, M.K, Noel, J.P, Moore, B.S. | | Deposit date: | 2012-12-11 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Stereospecific Vanadium-Dependent Haloperoxidase Family Enzymes in Napyradiomycin Biosynthesis.
Biochemistry, 2022
|
|
