6GX8
 
 | | Alpha-galactosidase from Thermotoga maritima in complex with hydrolysed cyclohexene-based carbasugar mimic of galactose | | Descriptor: | (1~{S},2~{S},3~{S},4~{S})-3-fluoranyl-6-(hydroxymethyl)cyclohex-5-ene-1,2,4-triol, Alpha-galactosidase, GLYCEROL, ... | | Authors: | Gloster, T.M, Oehler, V. | | Deposit date: | 2018-06-26 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Revealing the mechanism for covalent inhibition of glycoside hydrolases by carbasugars at an atomic level.
Nat Commun, 9, 2018
|
|
7O4O
 
 | | Structure of Staphylococcus aureus m1A22-tRNA methyltransferase in complex with S-adenosylhomocysteine | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Gloster, T.M, Czekster, C.M, da Silva, R.G. | | Deposit date: | 2021-04-06 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure, dynamics, and molecular inhibition of the Staphylococcus aureus m 1 A22-tRNA methyltransferase TrmK.
J.Biol.Chem., 298, 2022
|
|
7O4N
 
 | | Structure of Staphylococcus aureus m1A22-tRNA methyltransferase in complex with S-adenosylmethionine | | Descriptor: | GLYCEROL, S-ADENOSYLMETHIONINE, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Gloster, T.M, Czekster, C.M, da Silva, R.G. | | Deposit date: | 2021-04-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, dynamics, and molecular inhibition of the Staphylococcus aureus m 1 A22-tRNA methyltransferase TrmK.
J.Biol.Chem., 298, 2022
|
|
7O4M
 
 | | Structure of Staphylococcus aureus m1A22-tRNA methyltransferase | | Descriptor: | CITRIC ACID, GLYCEROL, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Gloster, T.M, Czekster, C.M, da Silva, R.G. | | Deposit date: | 2021-04-06 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure, dynamics, and molecular inhibition of the Staphylococcus aureus m 1 A22-tRNA methyltransferase TrmK.
J.Biol.Chem., 298, 2022
|
|
7P1V
 
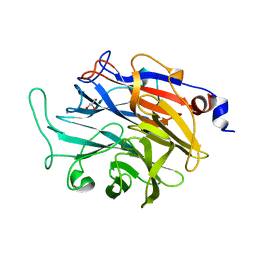 | | Apo structure of KDNase from Trichophyton Rubrum | | Descriptor: | CALCIUM ION, Extracellular sialidase/neuraminidase, GLYCEROL | | Authors: | Gloster, T.M, McMahon, S.A. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
7P1B
 
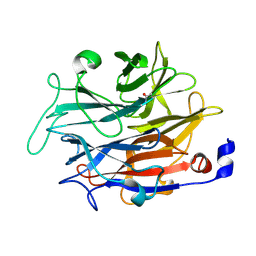 | |
7P1E
 
 | | Structure of KDNase from Aspergillus Terrerus in complex with 2,3-difluoro-2-keto-3-deoxynononic acid | | Descriptor: | (2R,3R,4R,5R,6S)-2,3-bis(fluoranyl)-4,5-bis(oxidanyl)-6-[(1R,2R)-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, 3-deoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, CALCIUM ION, ... | | Authors: | Gloster, T.M, McMahon, S.A. | | Deposit date: | 2021-07-01 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
7P1O
 
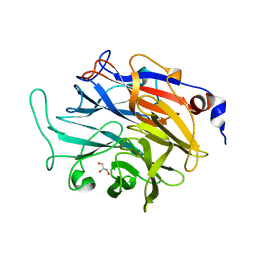 | |
7P1S
 
 | | Structure of KDNase from Trichophyton Rubrum in complex with 2,3-didehydro-2,3-dideoxy-D-glycero-D-galacto-nonulosonic acid. | | Descriptor: | 2,6-anhydro-3-deoxy-D-glycero-D-galacto-non-2-enonic acid, Extracellular sialidase/neuraminidase, SODIUM ION | | Authors: | Gloster, T.M, McMahon, S.A. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
7P1Q
 
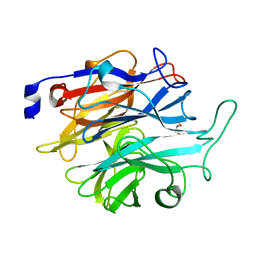 | |
7P1R
 
 | |
7P1D
 
 | |
7P1U
 
 | |
7P1F
 
 | | Structure of KDNase from Aspergillus terrerus in complex with 2,3-didehydro-2,3-dideoxy-D-glycero-D-galacto-nonulosonic acid. | | Descriptor: | 2,6-anhydro-3-deoxy-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Gloster, T.M, McMahon, S.A. | | Deposit date: | 2021-07-01 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Kinetic and Structural Characterization of Sialidases (Kdnases) from Ascomycete Fungal Pathogens.
Acs Chem.Biol., 16, 2021
|
|
2V38
 
 | | Family 5 endoglucanase Cel5A from Bacillus agaradhaerens in complex with cellobio-derived noeuromycin | | Descriptor: | (3S,4R,5R)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)PIPERIDIN-2-ONE, ENDOGLUCANASE 5A, GLYCEROL, ... | | Authors: | Gloster, T.M, Meloncelli, P.J, Money, V.A, Tarling, C.A, Davies, G.J, Withers, S.G, Stick, R.V. | | Deposit date: | 2007-06-13 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | D-Glucosylated Derivatives of Isofagomine and Noeuromycin and Their Potential as Inhibitors of Beta-Glycoside Hydrolases
To be Published
|
|
2WYN
 
 | | Structure of family 37 trehalase from Escherichia coli in complex with a casuarine-6-O-a-D-glucoside analogue | | Descriptor: | (1R,2R,3R,6R,7R,7AR)-3,7-BIS(HYDROXYMETHYL)HEXAHYDRO-1H-PYRROLIZINE-1,2,6-TRIOL, CALCIUM ION, PERIPLASMIC TREHALASE, ... | | Authors: | Gloster, T.M, Roberts, S.M, Davies, G.J, Cardona, F, Goti, A, Parmeggiani, C, Parenti, P, Fusi, P, Forcella, M, Cipolla, L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Casuarine-6-O-alpha-D-glucoside and its analogues are tight binding inhibitors of insect and bacterial trehalases.
Chem.Commun.(Camb.), 46, 2010
|
|
2W1W
 
 | | Native structure of a family 35 carbohydrate binding module from Clostridium thermocellum | | Descriptor: | CALCIUM ION, GLYCEROL, LIPOLYTIC ENZYME, ... | | Authors: | Gloster, T.M, Davies, G.J, Correia, M, Prates, J, Fontes, C, Gilbert, H.J. | | Deposit date: | 2008-10-21 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Evidence that Family 35 Carbohydrate Binding Modules Display Conserved Specificity But Divergent Function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WZ8
 
 | | Family 35 carbohydrate binding module from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CELLULOSOME PROTEIN DOCKERIN TYPE I, MAGNESIUM ION | | Authors: | Gloster, T.M, Davies, G.J, Fontes, C.M, Gilbert, H.J. | | Deposit date: | 2009-11-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Signature Active Site Architectures Illuminate the Molecular Basis for Ligand Specificity in Family 35 Carbohydrate Binding Module .
Biochemistry, 49, 2010
|
|
1UZ1
 
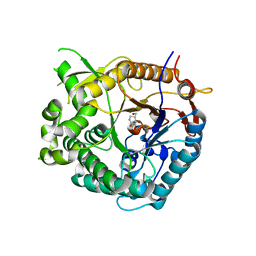 | | Family 1 b-glucosidase from Thermotoga maritima in complex with isofagomine lactam | | Descriptor: | (3S,4R,5R)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)PIPERIDIN-2-ONE, BETA-GLUCOSIDASE A | | Authors: | Gloster, T.M, Macdonald, J, Stick, R.V, Davies, G.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Common Inhibition of Both -Glucosidases and -Mannosidases by Isofagomine Lactam Reflects Different Conformational Itineraries for Pyranoside Hydrolysis
Chembiochem, 5, 2004
|
|
2J13
 
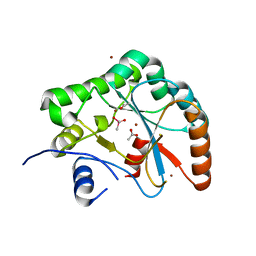 | | Structure of a family 4 carbohydrate esterase from Bacillus anthracis | | Descriptor: | ACETATE ION, CACODYLATE ION, POLYSACCHARIDE DEACETYLASE, ... | | Authors: | Gloster, T.M, Oberbarnscheidt, L, Taylor, E.J, Davies, G.J. | | Deposit date: | 2006-08-08 | | Release date: | 2006-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a Carbohydrate Esterase from Bacillus Anthracis.
Proteins: Struct., Funct., Bioinf., 66, 2007
|
|
2JJB
 
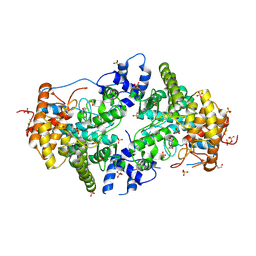 | | Family 37 trehalase from Escherichia coli in complex with casuarine-6- O-alpha-glucopyranose | | Descriptor: | 1,2-ETHANEDIOL, CASUARINE, PERIPLASMIC TREHALASE, ... | | Authors: | Gloster, T.M, Roberts, S, Davies, G.J, Cardona, F, Parmeggiani, C, Bonaccini, C, Gratteri, P, Sim, L, Rose, D.R, Goti, A. | | Deposit date: | 2008-03-28 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Total Syntheses of Casuarine and its 6-O-Alpha-Glucoside: Complementary Inhibition Towards Glycoside Hydrolases of the Gh31 and Gh37 Families.
Chemistry, 15, 2009
|
|
6YUD
 
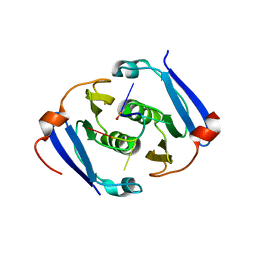 | | Structure of Csx3/Crn3 from Archaeoglobus fulgidus in complex with cyclic tetra-adenylate (cA4) | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), Uncharacterized protein AF_1864 | | Authors: | McQuarrie, S, Gloster, T.M, White, M.F, Graham, S, Athukoralage, J.S, Gruschow, S. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Tetramerisation of the CRISPR ring nuclease Crn3/Csx3 facilitates cyclic oligoadenylate cleavage.
Elife, 9, 2020
|
|
8PE3
 
 | | Structure of Csm6' from Streptococcus thermophilus in complex with cyclic hexa-adenylate (cA6) | | Descriptor: | CRISPR system endoribonuclease Csm6', Cyclic hexaadenosine monophosphate (cA6), RNA | | Authors: | McQuarrie, S.J, Athukoralage, J.S, McMahon, S.A, Graham, S, Ackerman, K, Bode, B.E, White, M.F, Gloster, T.M. | | Deposit date: | 2023-06-13 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Activation of Csm6 ribonuclease by cyclic nucleotide binding: in an emergency, twist to open.
Nucleic Acids Res., 51, 2023
|
|
8PCW
 
 | | Structure of Csm6' from Streptococcus thermophilus | | Descriptor: | CRISPR system endoribonuclease Csm6' | | Authors: | McQuarrie, S.J, Athukoralage, J.S, McMahon, S.A, Graham, S, Ackerman, K, Bode, B.E, White, M.F, Gloster, T.M. | | Deposit date: | 2023-06-11 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | Activation of Csm6 ribonuclease by cyclic nucleotide binding: in an emergency, twist to open.
Nucleic Acids Res., 51, 2023
|
|
6SCE
 
 | | Structure of a Type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate | | Descriptor: | Uncharacterized protein, cyclic oligoadenylate | | Authors: | McMahon, S.A, Zhu, W, Graham, S, White, M.F, Gloster, T.M. | | Deposit date: | 2019-07-24 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and mechanism of a Type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate.
Nat Commun, 11, 2020
|
|
