6KGC
 
 | | Crystal structure of CaDoc0917(R49D)-CaCohA2 complex at pH 5.4 | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION, Probably cellulosomal scaffolding protein, ... | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
6KGD
 
 | | Crystal structure of CaDoc0917(R49D)-CaCohA2 complex at pH 8.0 | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION, Probably cellulosomal scaffolding protein, ... | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
6KGF
 
 | | Crystal structure of CaDoc0917(R16D)-CaCohA2 complex at pH 8.2 | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION, Probably cellulosomal scaffolding protein, ... | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
6KGE
 
 | | Crystal structure of CaDoc0917(R16D)-CaCohA2 complex at pH 5.5 | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION, Probably cellulosomal scaffolding protein, ... | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
6KG8
 
 | | Solution structure of CaCohA2 from Clostridium acetobutylicum | | Descriptor: | Probably cellulosomal scaffolding protein, secreted cellulose-binding and cohesin domain | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
3OMG
 
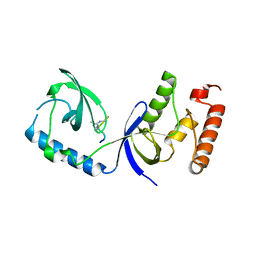 | | Structure of human SND1 extended tudor domain in complex with the symmetrically dimethylated arginine PIWIL1 peptide R14me2s | | Descriptor: | Staphylococcal nuclease domain-containing protein 1, dimethylated arginine peptide R14me2s | | Authors: | Lam, R, Liu, K, Guo, Y.H, Bian, C.B, Xu, C, MacKenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5I05
 
 | |
8YZ7
 
 | |
8XYC
 
 | | Ternary structure of dVemCas12e-sgRNA-dsDNA | | Descriptor: | DNA (35-MER), RNA (147-MER), dVemCas12e | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2024-01-19 | | Release date: | 2024-10-23 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Cas12e orthologs evolve variable structural elements to facilitate dsDNA cleavage.
Nat Commun, 15, 2024
|
|
8Y2I
 
 | |
7JT4
 
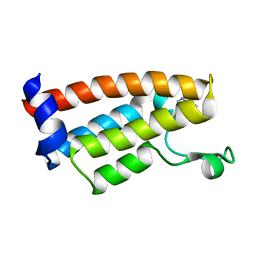 | | Crystal Structure of BPTF bromodomain labelled with 5-fluoro-tryptophan | | Descriptor: | Nucleosome-remodeling factor subunit BPTF | | Authors: | Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2020-08-17 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Selectivity, ligand deconstruction, and cellular activity analysis of a BPTF bromodomain inhibitor
Org.Biomol.Chem., 17, 2019
|
|
6E5B
 
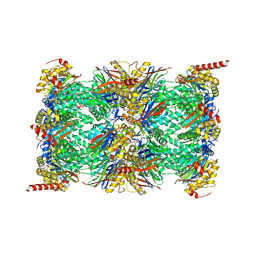 | | Human Immunoproteasome 20S particle in complex with compound 1 | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Steinbacher, S, Augustin, M, Blaesse, M, Harris, S.F. | | Deposit date: | 2018-07-19 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Design and Evaluation of Highly Selective Human Immunoproteasome Inhibitors Reveal a Compensatory Process That Preserves Immune Cell Viability.
J.Med.Chem., 62, 2019
|
|
7V59
 
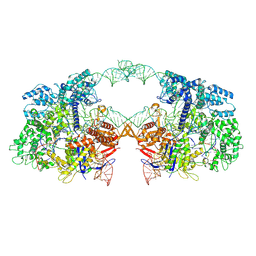 | | Cryo-EM structure of spyCas9-sgRNA-DNA dimer | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (49-MER), RNA (115-MER) | | Authors: | Liu, J, Deng, P. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (5.26 Å) | | Cite: | Nonspecific interactions between SpCas9 and dsDNA sites located downstream of the PAM mediate facilitated diffusion to accelerate target search.
Chem Sci, 12, 2021
|
|
7WKU
 
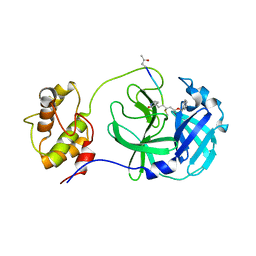 | | Structure of PDCoV Mpro in complex with an inhibitor | | Descriptor: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, Peptidase C30 | | Authors: | Wang, F.H, Yang, H.T. | | Deposit date: | 2022-01-11 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure of the Porcine Deltacoronavirus Main Protease Reveals a Conserved Target for the Design of Antivirals.
Viruses, 14, 2022
|
|
7Y1T
 
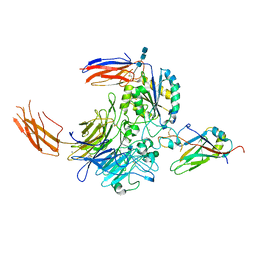 | | Complex of integrin alphaV/beta8 and L-TGF-beta1 at a ratio of 1:2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duan, Z, Zhang, Z. | | Deposit date: | 2022-06-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Specificity of TGF-beta 1 signal designated by LRRC33 and integrin alpha V beta 8.
Nat Commun, 13, 2022
|
|
7Y1R
 
 | | Human L-TGF-beta1 in complex with the anchor protein LRRC33 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duan, Z, Zhang, Z. | | Deposit date: | 2022-06-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Specificity of TGF-beta 1 signal designated by LRRC33 and integrin alpha V beta 8.
Nat Commun, 13, 2022
|
|
7CKF
 
 | | The N-terminus of interferon-inducible antiviral protein-dimer | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, Guanylate-binding protein 5, ... | | Authors: | Cui, W, Yang, H.T. | | Deposit date: | 2020-07-16 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.284 Å) | | Cite: | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CG5
 
 | |
7CG8
 
 | | Structure of the sensor domain (short construct) of the anti-sigma factor RsgI4 in Pseudobacteroides cellulosolvens | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, ACETATE ION, Anti-sigma factor RsgI, ... | | Authors: | Dong, S, Feng, Y. | | Deposit date: | 2020-06-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the sensor domain of the anti-sigma factor RsgI4 in Pseudobacteroides cellulosolvens
To Be Published
|
|
7CG1
 
 | |
7D5L
 
 | | Discovery of BMS-986144, a Third Generation, Pan Genotype NS3/4A Protease Inhibitor for the Treatment of Hepatitis C Virus Infection | | Descriptor: | NS3/4A Protease, ZINC ION, [1,1,1-tris(fluoranyl)-2-methyl-propan-2-yl] ~{N}-[(1~{S},4~{R},6~{S},7~{Z},11~{R},13~{R},14~{S},18~{R})-13-ethyl-18-(7-fluoranyl-6-methoxy-isoquinolin-1-yl)oxy-11-methyl-4-[(1-methylcyclopropyl)sulfonylcarbamoyl]-2,15-bis(oxidanylidene)-3,16-diazatricyclo[14.3.0.0^{4,6}]nonadec-7-en-14-yl]carbamate | | Authors: | Ghosh, K, Anumula, R, Kumar, A. | | Deposit date: | 2020-09-26 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of BMS-986144, a Third-Generation, Pan-Genotype NS3/4A Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 63, 2020
|
|
7CCE
 
 | |
7EJE
 
 | | human RAD51 post-synaptic complex | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Zhao, L.Y, Xu, J.F, Wang, H.W. | | Deposit date: | 2021-04-02 | | Release date: | 2022-03-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Mechanisms of distinctive mismatch tolerance between Rad51 and Dmc1 in homologous recombination.
Nucleic Acids Res., 49, 2021
|
|
7EJC
 
 | | human RAD51 presynaptic complex | | Descriptor: | 4-bromanyl-N-(4-bromophenyl)-3-[(phenylmethyl)sulfamoyl]benzamide, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Zhao, L.Y, Xu, J.F, Wang, H.W. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Mechanisms of distinctive mismatch tolerance between Rad51 and Dmc1 in homologous recombination.
Nucleic Acids Res., 49, 2021
|
|
7EJ6
 
 | | Yeast Dmc1 presynaptic complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), HLJ1_G0016300.mRNA.1.CDS.1, ... | | Authors: | Zhao, L.Y, Xu, J.F, Wang, H.W. | | Deposit date: | 2021-04-01 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Mechanisms of distinctive mismatch tolerance between Rad51 and Dmc1 in homologous recombination.
Nucleic Acids Res., 49, 2021
|
|
