4DJX
 
 | |
6A7A
 
 | | AKR1C1 complexed with new inhibitor with novel scaffold | | Descriptor: | (4R)-6-amino-4-(4-hydroxy-3-methoxy-5-nitrophenyl)-3-propyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X, Zhao, Y, Zhang, H, Chen, Y. | | Deposit date: | 2018-07-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
6A7B
 
 | | AKR1C3 complexed with new inhibitor with novel scaffold | | Descriptor: | (4R)-6-amino-4-(4-hydroxy-3-methoxy-5-nitrophenyl)-3-propyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, Aldo-keto reductase family 1 member C3, DIMETHYLFORMAMIDE, ... | | Authors: | Zheng, X, Zhao, Y, Zhang, H, Chen, Y. | | Deposit date: | 2018-07-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
4QGE
 
 | | phosphodiesterase-9A in complex with inhibitor WYQ-C36D | | Descriptor: | MAGNESIUM ION, N~2~-(1-cyclopentyl-4-oxo-4,7-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl)-N-(4-methoxyphenyl)-D-alaninamide, Phosphodiesterase 9A, ... | | Authors: | Shao, Y.-X, Huang, M, Cui, W, Ke, H. | | Deposit date: | 2014-05-22 | | Release date: | 2014-12-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a Phosphodiesterase 9A Inhibitor as a Potential Hypoglycemic Agent.
J.Med.Chem., 57, 2014
|
|
4DJY
 
 | |
3EUV
 
 | | Crystal structure of FTase(ALPHA-subunit; BETA-subunit DELTA C10, W102T, Y154T) in complex with BiotinGPP | | Descriptor: | (2E,6E)-3,7-dimethyl-8-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)octa-2,6-dien-1-yl trihydrogen diphosphate, Protein farnesyltransferase subunit beta, Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha, ... | | Authors: | Guo, Z, Nguyen, U.T.T, Delon, C, Bon, R.S, Blankenfeldt, W, Goody, R.S, Waldmann, H, Wolters, D, Alexandrov, K. | | Deposit date: | 2008-10-11 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Analysis of the eukaryotic prenylome by isoprenoid affinity tagging
Nat.Chem.Biol., 5, 2009
|
|
3EU5
 
 | | Crystal structure of FTase(ALPHA-subunit; BETA-subunit DELTA C10) in complex with BiotinGPP | | Descriptor: | (2E,6E)-3,7-dimethyl-8-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)octa-2,6-dien-1-yl trihydrogen diphosphate, Protein farnesyltransferase subunit beta, Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha, ... | | Authors: | Guo, Z, Nguyen, U.T.T, Delon, C, Bon, R.S, Blankenfeldt, W, Goody, R.S, Waldmann, H, Wolters, D, Alexandrov, K. | | Deposit date: | 2008-10-09 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the eukaryotic prenylome by isoprenoid affinity tagging
Nat.Chem.Biol., 5, 2009
|
|
8TN1
 
 | |
8TNB
 
 | |
8TNC
 
 | |
8TND
 
 | |
8TN6
 
 | |
3G6J
 
 | | C3b in complex with a C3b specific Fab | | Descriptor: | CALCIUM ION, Complement C3 alpha chain, Complement C3 beta chain, ... | | Authors: | Wiesmann, C. | | Deposit date: | 2009-02-06 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Functional Analysis of a C3b-specific Antibody That Selectively Inhibits the Alternative Pathway of Complement
J.Biol.Chem., 284, 2009
|
|
8F2C
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML3006a | | Descriptor: | (1R,2S,5S)-N-[(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-(2-oxopyrrolidin-1-yl)butan-2-yl]-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-11-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
8F02
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML2006a4 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(3,3-dimethylazetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-11-01 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
8EZV
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML2006a | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-11-01 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
8EZZ
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML2006a2 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(3,3-difluoroazetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-11-01 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
8F2D
 
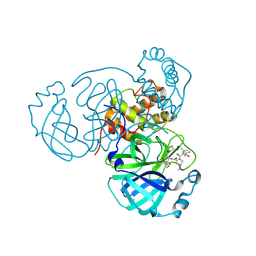 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML4006a | | Descriptor: | (1R,2S,5S)-N-[(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-(2-oxopiperidin-1-yl)butan-2-yl]-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-11-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
8GY1
 
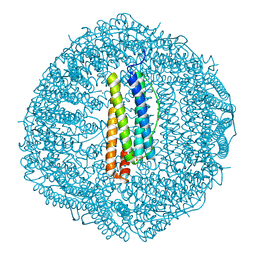 | |
5VLH
 
 | | Short PCSK9 delta-P' complex with peptide Pep1 | | Descriptor: | ACE-THR-VAL-PHE-THR-SER-TRP-GLU-GLU-TYR-LEU-ASP-TRP-VAL-NH2, CALCIUM ION, CYS-ARG-LEU-PRO-TRP-ASN-LEU-GLN-ARG-ILE-GLY-LEU-PRO-CYS, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5VLL
 
 | | Short PCSK9 delta-P' complex with peptide Pep3 | | Descriptor: | ACE-THR-VAL-PHE-THR-SER-TRP-GLU-GLU-TYR-LEU-ASP-TRP-VAL-NH2, CALCIUM ION, CYS-PHE-ILE-PRO-TRP-ASN-LEU-GLN-ARG-ILE-GLY-LEU-LEU-CYS, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5VL7
 
 | | PCSK9 complex with Fab33 | | Descriptor: | Fab33 heavy chain, Fab33 light chain, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
8HL1
 
 | |
5VLA
 
 | | Short PCSK9 delta-P' complex with Fusion2 peptide | | Descriptor: | CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, THR-VAL-PHE-THR-SER-TRP-GLU-GLU-TYR-LEU-ASP-TRP-VAL-MET-PRO-TRP-ASN-LEU-VAL-ARG-ILE-GLY-LEU-LEU | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5VLK
 
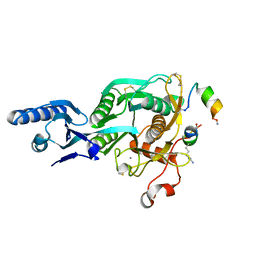 | |
