8QTT
 
 | |
8QTF
 
 | |
6C8Z
 
 | | Last common ancestor of ADP-dependent phosphofructokinases from Methanosarcinales | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADP-dependent phosphofructokinase, MAGNESIUM ION, ... | | Authors: | Castro-Fernandez, V, Gonzalez-Ordenes, F, Munoz, S, Fuentes, N, Leonardo, D, Fuentealba, M, Herrera-Morande, A, Maturana, P, Villalobos, P, Garratt, R. | | Deposit date: | 2018-01-25 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | ADP-Dependent Kinases From the Archaeal OrderMethanosarcinalesAdapt to Salt by a Non-canonical Evolutionarily Conserved Strategy.
Front Microbiol, 9, 2018
|
|
6RUX
 
 | | P46, an immunodominant surface protein from Mycoplasma hyopneumoniae | | Descriptor: | 46 kDa surface antigen, SODIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Guasch, A, Gonzalez-Gonzalez, L, Fita, I. | | Deposit date: | 2019-05-29 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of P46, an immunodominant surface protein from Mycoplasma hyopneumoniae: interaction with a monoclonal antibody.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6S3T
 
 | | P46, an immunodominant surface protein from Mycoplasma hyopneumoniae | | Descriptor: | 46 kDa surface antigen, Immunoglobulin heavy chain, Immunoglobulin light chain, ... | | Authors: | Guasch, A, Gonzalez-Gonzalez, L, Fita, I. | | Deposit date: | 2019-06-26 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of P46, an immunodominant surface protein from Mycoplasma hyopneumoniae: interaction with a monoclonal antibody.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5C26
 
 | | Crystal structure of SYK in complex with compound 1 | | Descriptor: | 3-{8-[(3,4-dimethoxyphenyl)amino]imidazo[1,2-a]pyrazin-6-yl}benzamide, GLU-VAL-PTR-GLU-SER-PRO, Tyrosine-protein kinase SYK | | Authors: | Han, S, Chang, J. | | Deposit date: | 2015-06-15 | | Release date: | 2015-10-07 | | Last modified: | 2016-02-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Imidazotriazines: Spleen Tyrosine Kinase (Syk) Inhibitors Identified by Free-Energy Perturbation (FEP).
Chemmedchem, 11, 2016
|
|
1DX9
 
 | | W57A Apoflavodoxin from Anabaena | | Descriptor: | Flavodoxin, SULFATE ION | | Authors: | Romero, A, Sancho, J. | | Deposit date: | 1999-12-23 | | Release date: | 2000-04-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dissecting the Energetics of the Apoflavodoxin-Fmn Complex
J.Biol.Chem., 275, 2000
|
|
5C27
 
 | | Crystal structure of SYK in complex with compound 2 | | Descriptor: | 3-{8-[(3,4-dimethoxyphenyl)amino]imidazo[1,2-a]pyrazin-6-yl}-N-[4-(methylcarbamoyl)phenyl]benzamide, GLU-VAL-TYR-GLU-SER, GLYCEROL, ... | | Authors: | Han, S, Chang, J. | | Deposit date: | 2015-06-15 | | Release date: | 2015-10-07 | | Last modified: | 2016-02-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Imidazotriazines: Spleen Tyrosine Kinase (Syk) Inhibitors Identified by Free-Energy Perturbation (FEP).
Chemmedchem, 11, 2016
|
|
1TCD
 
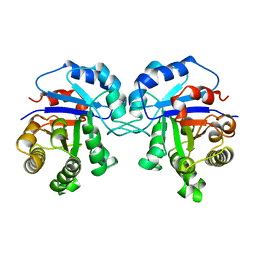 | | TRYPANOSOMA CRUZI TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Maldonado, E, Soriano-Garcia, M, Cabrera, N, Garza-Ramos, G, Tuena De Gomez-Puyou, M, Gomez-Puyou, A, Perez-Montfort, R. | | Deposit date: | 1998-01-29 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Differences in the intersubunit contacts in triosephosphate isomerase from two closely related pathogenic trypanosomes.
J.Mol.Biol., 283, 1998
|
|
6ENV
 
 | |
6ENW
 
 | |
6FX8
 
 | |
6FX9
 
 | |
6DXG
 
 | | amidobenzimidazole (ABZI) STING agonists | | Descriptor: | 2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-1-[(2R)-2-hydroxy-2-phenylethyl]-1H-benzimidazole-5-carboxamide, CALCIUM ION, Stimulator of interferon protein | | Authors: | Concha, N.O. | | Deposit date: | 2018-06-28 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Design of amidobenzimidazole STING receptor agonists with systemic activity.
Nature, 564, 2018
|
|
6DXL
 
 | | Linked amidobenzimidazole STING agonist | | Descriptor: | 1,1'-(butane-1,4-diyl)bis{2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-1H-benzimidazole-5-carboxamide}, CALCIUM ION, Stimulator of interferon protein | | Authors: | Concha, N.O. | | Deposit date: | 2018-06-29 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design of amidobenzimidazole STING receptor agonists with systemic activity.
Nature, 564, 2018
|
|
6EQG
 
 | | Crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis in spacegroup P21 | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EQH
 
 | | Crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis in spacegroup C2221 | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EQE
 
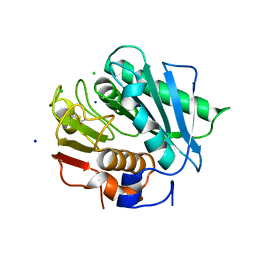 | | High resolution crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SODIUM ION | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EQF
 
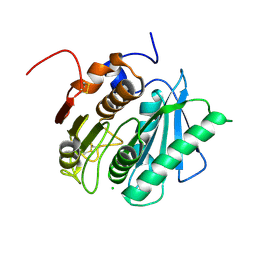 | | Crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis in spacegroup P212121 | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EQD
 
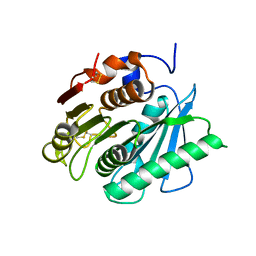 | | Crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis collected at long wavelength | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2018-05-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1CI1
 
 | | CRYSTAL STRUCTURE OF TRIOSEPHOSPHATE ISOMERASE FROM TRYPANOSOMA CRUZI IN HEXANE | | Descriptor: | HEXANE, PROTEIN (TRIOSEPHOSPHATE ISOMERASE) | | Authors: | Gao, X.-G, Maldondo, E, Perez-Montfort, R, De Gomez-Puyou, M.T, Gomez-Puyou, A, Rodriguez-Romero, A. | | Deposit date: | 1999-04-06 | | Release date: | 1999-09-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of triosephosphate isomerase from Trypanosoma cruzi in hexane.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3IQ5
 
 | |
3IQ6
 
 | |
1O50
 
 | |
1O5H
 
 | |
