7B8X
 
 | | Notum-Fragment 210 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-13 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BAP
 
 | | Notum Fragment 648 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-16 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BCK
 
 | | Notum Fragment 791 | | Descriptor: | (2S)-N-(4-Methoxybenzyl)tetrahydrofuran-2-carboxamide, Palmitoleoyl-protein carboxylesterase NOTUM, SULFATE ION | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-19 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BD9
 
 | | Notum Fragment 886 | | Descriptor: | (4-chloranyl-2-methyl-pyrazol-3-yl)-piperidin-1-yl-methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
4LZV
 
 | |
5T5T
 
 | | AMPK bound to allosteric activator | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2016-08-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Selective Activation of AMPK beta 1-Containing Isoforms Improves Kidney Function in a Rat Model of Diabetic Nephropathy.
J. Pharmacol. Exp. Ther., 361, 2017
|
|
7JRN
 
 | | Crystal structure of the wild type SARS-CoV-2 papain-like protease (PLPro) with inhibitor GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, SULFATE ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-08-12 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of SARS-CoV-2 Papain-like Protease Inhibitors through a Combination of High-Throughput Screening and a FlipGFP-Based Reporter Assay.
Acs Cent.Sci., 7, 2021
|
|
4N1U
 
 | | Structure of human MTH1 in complex with TH588 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N~4~-cyclopropyl-6-(2,3-dichlorophenyl)pyrimidine-2,4-diamine, SULFATE ION | | Authors: | Berntsson, R.P.-A, Jemth, A, Gustafsson, R, Svensson, L.M, Helleday, T, Stenmark, P. | | Deposit date: | 2013-10-04 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool.
Nature, 508, 2014
|
|
5GW8
 
 | | Crystal structure of a putative DAG-like lipase (MgMDL2) from Malassezia globosa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Xu, J, Xu, H, Liu, J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Malassezia globosa MgMDL2 lipase: Crystal structure and rational modification of substrate specificity.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
7B99
 
 | | Notum Fragment 283 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B8U
 
 | | Notum-Fragment 201 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(piperidin-1-yl)-1,2,5-oxadiazol-3-amine, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-13 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B9I
 
 | | Notum Fragment 297 | | Descriptor: | 1-(1,3-benzodioxol-5-yl)-~{N}-(pyridin-2-ylmethyl)methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BA1
 
 | | Notum Fragment 634 | | Descriptor: | 1-ethanoylpiperidine-4-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-15 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B9N
 
 | | Notum Fragment 588 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BC9
 
 | | Notum Fragment 690 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-methoxy-N-(4-phenyl-1,3-thiazol-2-yl)acetamide, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-18 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BD3
 
 | | Notum Fragment 823 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-methoxy-6-phenyl-pyrimidin-2-amine, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7BCC
 
 | | Notum Fragment 705 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(4-pyrrol-1-ylphenyl)morpholine, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-12-19 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
4LZU
 
 | |
4MI5
 
 | | Crystal structure of the EZH2 SET domain | | Descriptor: | Histone-lysine N-methyltransferase EZH2, SULFATE ION, ZINC ION | | Authors: | Antonysamy, S, Condon, B, Druzina, Z, Bonanno, J, Gheyi, T, Macewan, I, Zhang, A, Ashok, S, Russell, M, Luz, J.G. | | Deposit date: | 2013-08-30 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Context of Disease-Associated Mutations and Putative Mechanism of Autoinhibition Revealed by X-Ray Crystallographic Analysis of the EZH2-SET Domain.
Plos One, 8, 2013
|
|
4N1T
 
 | | Structure of human MTH1 in complex with TH287 | | Descriptor: | 6-(2,3-dichlorophenyl)-N~4~-methylpyrimidine-2,4-diamine, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Berntsson, R.P.-A, Jemth, A, Gustafsson, R, Svensson, L.M, Helleday, T, Stenmark, P. | | Deposit date: | 2013-10-04 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool.
Nature, 508, 2014
|
|
4KMX
 
 | | Human folate receptor alpha (FOLR1) at acidic pH, hexagonal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wibowo, A.S, Dann III, C.E. | | Deposit date: | 2013-05-08 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of human folate receptors reveal biological trafficking states and diversity in folate and antifolate recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5Z72
 
 | |
4D9N
 
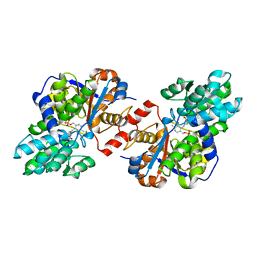 | | Crystal structure of Diaminopropionate ammonia lyase from Escherichia coli in complex with D-serine | | Descriptor: | D-SERINE, Diaminopropionate ammonia-lyase | | Authors: | Bisht, S, Rajaram, V, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Escherichia coli Diaminopropionate Ammonia-lyase Reveals Mechanism of Enzyme Activation and Catalysis
J.Biol.Chem., 287, 2012
|
|
5IKI
 
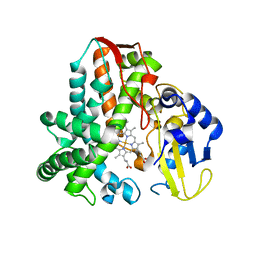 | | CYP106A2 WITH SUBSTRATE ABIETIC ACID | | Descriptor: | Abietic acid, Cytochrome P450(MEG), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Janocha, S, Carius, Y, Bernhardt, R, Lancaster, C.R.D. | | Deposit date: | 2016-03-03 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of CYP106A2 in Substrate-Free and Substrate-Bound Form.
Chembiochem, 17, 2016
|
|
2V8L
 
 | |
