7X0Q
 
 | |
7X0R
 
 | |
7X0I
 
 | |
7X0N
 
 | |
7X0J
 
 | |
7W06
 
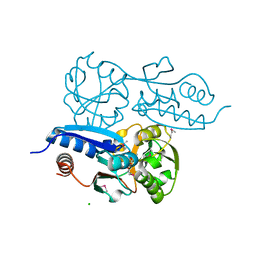 | | Itaconate inducible LysR-Type Transcriptional regulator (ITCR) in complex with itaconate (SeMet labeled), Space group C121. | | Descriptor: | 2-methylidenebutanedioic acid, CHLORIDE ION, SULFATE ION, ... | | Authors: | Sun, P.K, Wang, B, Wang, Z.X, Qi, S, Li, X.J. | | Deposit date: | 2021-11-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A genetically encoded fluorescent biosensor for detecting itaconate with subcellular resolution in living macrophages.
Nat Commun, 13, 2022
|
|
7W07
 
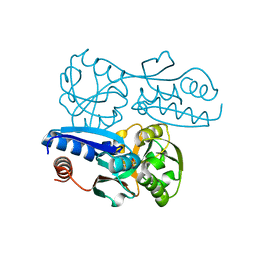 | | Itaconate inducible LysR-Type Transcriptional regulator (ITCR) in complex with itaconate, Space group C121. | | Descriptor: | 2-methylidenebutanedioic acid, SULFATE ION, Transcriptional regulator, ... | | Authors: | Sun, P.K, Wang, B, Li, X.J. | | Deposit date: | 2021-11-17 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | A genetically encoded fluorescent biosensor for detecting itaconate with subcellular resolution in living macrophages.
Nat Commun, 13, 2022
|
|
7W08
 
 | |
7X0M
 
 | |
7Y78
 
 | | Crystal structure of Cry78Aa | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, Toxin | | Authors: | Cao, B.B, Nie, Y.F, Wang, N.C, Guan, Z.Y, Zhang, D.L, Zhang, J. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of Cry78Aa from Bacillus thuringiensis provides insights into its insecticidal activity.
Commun Biol, 5, 2022
|
|
7Y79
 
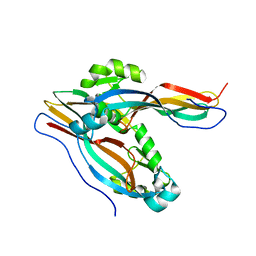 | | Crystal structure of Cry78Aa | | Descriptor: | Toxin | | Authors: | Cao, B.B, Nie, Y.F, Wang, N.C, Guan, Z.Y, Zhang, D.L, Zhang, J. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The crystal structure of Cry78Aa from Bacillus thuringiensis provides insights into its insecticidal activity.
Commun Biol, 5, 2022
|
|
8RIY
 
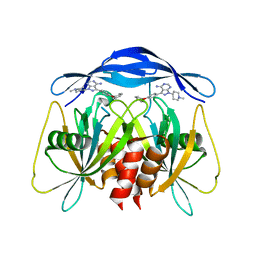 | | Human NUDT5 with ibrutinib derivative | | Descriptor: | 1-(1-methylpiperidin-4-yl)-3-(4-phenoxyphenyl)pyrazolo[3,4-d]pyrimidin-4-amine, ADP-sugar pyrophosphatase | | Authors: | Balikci-Akil, E, Elkins, J.M, Huber, K.V.M. | | Deposit date: | 2023-12-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Unexpected Noncovalent Off-Target Activity of Clinical BTK Inhibitors Leads to Discovery of a Dual NUDT5/14 Antagonist.
J.Med.Chem., 67, 2024
|
|
8RDZ
 
 | |
6IVU
 
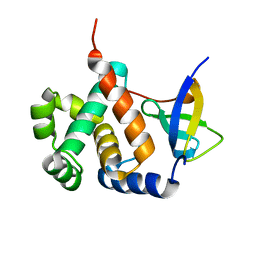 | |
6IVS
 
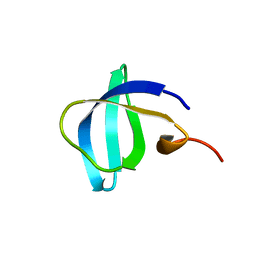 | |
3OMC
 
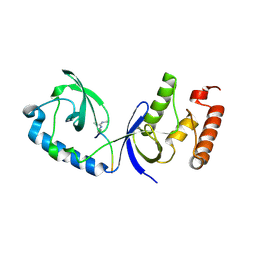 | | Structure of human SND1 extended tudor domain in complex with the symmetrically dimethylated arginine PIWIL1 peptide R4me2s | | Descriptor: | CHLORIDE ION, SYNTHETIC PEPTIDE, Staphylococcal nuclease domain-containing protein 1 | | Authors: | Lam, R, Liu, K, Guo, Y.H, Bian, C.B, Xu, C, MacKenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7CMA
 
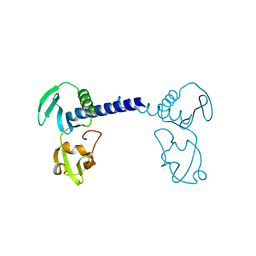 | | Structure of A151R from African swine fever virus Georgia | | Descriptor: | A151R, ZINC ION | | Authors: | Niu, D, Liu, K, Huang, J, Chen, C, Liu, W, Guo, R. | | Deposit date: | 2020-07-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure basis of non-structural protein pA151R from African Swine Fever Virus.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
8JIX
 
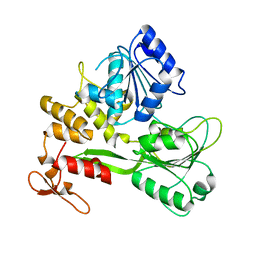 | | Crystal structure of the Bagaza virus helicase and structure-based discovery of a novel inhibitor | | Descriptor: | Genome polyprotein | | Authors: | Zhao, R, Shu, W, Cao, J.M, Zhou, X, Wang, D.P. | | Deposit date: | 2023-05-29 | | Release date: | 2024-03-06 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure-based discovery of dual-target inhibitors of the helicase from bagaza virus.
Int.J.Biol.Macromol., 294, 2025
|
|
7DCD
 
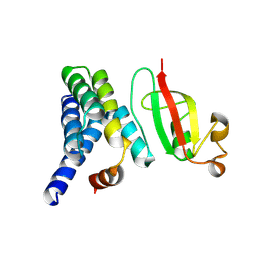 | |
6ISU
 
 | | Crystal structure of Lys27-linked di-ubiquitin in complex with its selective interacting protein UCHL3 | | Descriptor: | Ubiquitin, Ubiquitin carboxyl-terminal hydrolase isozyme L3 | | Authors: | Ding, S, Pan, M, Zheng, Q, Ren, Y, Hong, D. | | Deposit date: | 2018-11-19 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.866 Å) | | Cite: | Chemical Protein Synthesis Enabled Mechanistic Studies on the Molecular Recognition of K27-linked Ubiquitin Chains.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
7DD1
 
 | |
8K09
 
 | | Pq3-O-UGT2 with 20(S)-Ginsenoside F2 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-[[(3~{S},5~{R},8~{R},9~{R},10~{R},12~{R},13~{R},14~{R},17~{S})-17-[(2~{S})-2-[(2~{S},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-6-methyl-hept-5-en-2-yl]-4,4,8,10,14-pentamethyl-12-oxidanyl-2,3,5,6,7,9,11,12,13,15,16,17-dodecahydro-1~{H}-cyclopenta[a]phenanthren-3-yl]oxy]oxane-3,4,5-triol, Glycosyltransferase | | Authors: | Mei, K.R, Ji, Q.S, Liu, Y.R. | | Deposit date: | 2023-07-07 | | Release date: | 2025-01-29 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structural Insights into the Substrate Recognition of Ginsenoside Glycosyltransferase Pq3-O-UGT2.
Adv Sci, 12, 2025
|
|
8JZQ
 
 | |
8K08
 
 | | Pq3-O-UGT2 with 20(S)-Ginsenoside Rh2 | | Descriptor: | Ginsenoside Rh2, Glycosyltransferase | | Authors: | Ji, Q.S, Liu, Y.R, Mei, K.R. | | Deposit date: | 2023-07-07 | | Release date: | 2025-01-29 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Insights into the Substrate Recognition of Ginsenoside Glycosyltransferase Pq3-O-UGT2.
Adv Sci, 12, 2025
|
|
6KG9
 
 | | Solution structure of CaDoc0917 from Clostridium acetobutylicum | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
