8B4V
 
 | | X-ray structure of furin (PCSK3) in complex with benzamidine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BENZAMIDINE, CALCIUM ION, ... | | Authors: | Dahms, S.O, Brandstetter, H. | | Deposit date: | 2022-09-21 | | Release date: | 2023-10-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-Based Design, Synthesis, and Characterization of Aminoisoindole-Derived Furin Inhibitors.
Chemmedchem, 19, 2024
|
|
8CCI
 
 | | Crystal structure of Mycobacterium smegmatis thioredoxin reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fuesser, F.T, Koch, O, Kuemmel, D. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Novel starting points for fragment-based drug design against mycobacterial thioredoxin reductase identified using crystallographic fragment screening.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
4YHU
 
 | | Yeast Prp3 C-terminal fragment 296-469 | | Descriptor: | U4/U6 small nuclear ribonucleoprotein PRP3, YTTRIUM (III) ION | | Authors: | Liu, S, Wahl, M.C. | | Deposit date: | 2015-02-27 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A composite double-/single-stranded RNA-binding region in protein Prp3 supports tri-snRNP stability and splicing.
Elife, 4, 2015
|
|
4YHV
 
 | | Yeast Prp3 C-terminal fragment 325-469 | | Descriptor: | ACETIC ACID, U4/U6 small nuclear ribonucleoprotein PRP3 | | Authors: | Liu, S, Wahl, M.C. | | Deposit date: | 2015-02-27 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A composite double-/single-stranded RNA-binding region in protein Prp3 supports tri-snRNP stability and splicing.
Elife, 4, 2015
|
|
4YHW
 
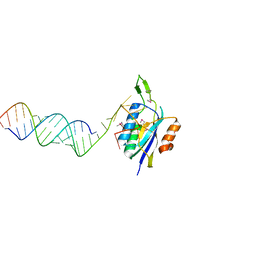 | | Yeast Prp3 (296-469) in complex with fragment of U4/U6 di-snRNA | | Descriptor: | U4 snRNA fragment, U4/U6 small nuclear ribonucleoprotein PRP3, U6 snRNA fragment | | Authors: | Liu, S, Wahl, M.C. | | Deposit date: | 2015-02-27 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | A composite double-/single-stranded RNA-binding region in protein Prp3 supports tri-snRNP stability and splicing.
Elife, 4, 2015
|
|
7AQE
 
 | | Structure of SARS-CoV-2 Main Protease bound to UNC-2327 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Meents, A. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
8B4W
 
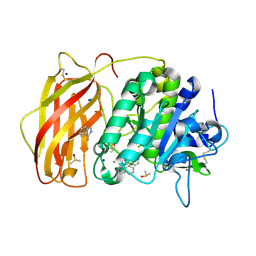 | | X-ray structure of furin (PCSK3) in complex with 1H-isoindol-3-amine | | Descriptor: | 1H-isoindol-3-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dahms, S.O, Brandstetter, H. | | Deposit date: | 2022-09-21 | | Release date: | 2023-10-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-Based Design, Synthesis, and Characterization of Aminoisoindole-Derived Furin Inhibitors.
Chemmedchem, 19, 2024
|
|
8B4X
 
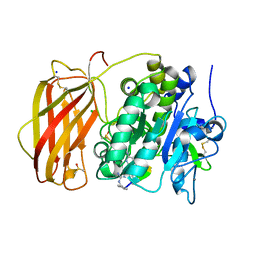 | | X-ray structure of furin (PCSK3) in complex with Guanidinomethyl-Phac-R-Tle-K-6-(aminomethyl)-3-amino-isoindol | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Dahms, S.O, Brandstetter, H. | | Deposit date: | 2022-09-21 | | Release date: | 2023-10-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-Based Design, Synthesis, and Characterization of Aminoisoindole-Derived Furin Inhibitors.
Chemmedchem, 19, 2024
|
|
8BM8
 
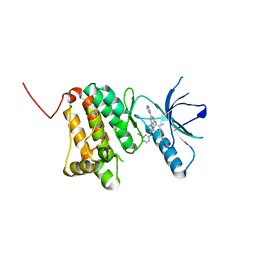 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 11 | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(3-cyanophenyl)-4-methyl-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-10 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | (placeholder, will be filled after publication)
To Be Published
|
|
8OYH
 
 | | X-ray structure of furin (PCSK3) in complex with Guanidinomethyl-Phac-Can-Tle-Can-6-(aminomethyl)-3-amino-isoindol | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Dahms, S.O, Brandstetter, H. | | Deposit date: | 2023-05-04 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-Based Design, Synthesis, and Characterization of Aminoisoindole-Derived Furin Inhibitors.
Chemmedchem, 19, 2024
|
|
8OGK
 
 | |
5ROQ
 
 | | PanDDA analysis group deposition -- Proteinase K changed state model for fragment Frag Xtal Screen A12a | | Descriptor: | (2R)-(3-chlorophenyl)(hydroxy)ethanoic acid, Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5ROJ
 
 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo59 | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5RPL
 
 | | PanDDA analysis group deposition -- Proteinase K changed state model for fragment Frag Xtal Screen C8a | | Descriptor: | (4-methoxycarbonylphenyl)methylazanium, Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5ROE
 
 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo6 | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5RPS
 
 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo60 | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5ROF
 
 | | PanDDA analysis group deposition -- Proteinase K changed state model for fragment Frag Xtal Screen C11a | | Descriptor: | 2-(1H-indol-3-yl)-N-[(1-methyl-1H-pyrrol-2-yl)methyl]ethanamine, Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5RPV
 
 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo35 | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5ROH
 
 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo61 | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5RPY
 
 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo10 | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5ROI
 
 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo30 | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5ROK
 
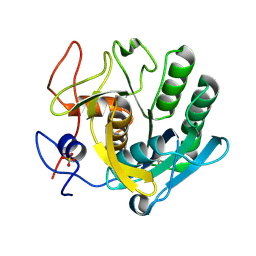 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo37 | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5ROO
 
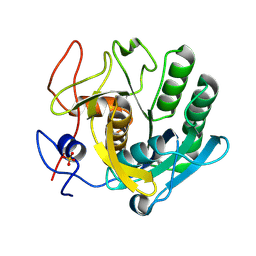 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo73 | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5ROP
 
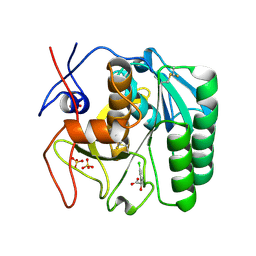 | | PanDDA analysis group deposition -- Proteinase K changed state model for fragment Frag Xtal Screen A12a at Room Temperature | | Descriptor: | (2R)-(3-chlorophenyl)(hydroxy)ethanoic acid, Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5RPT
 
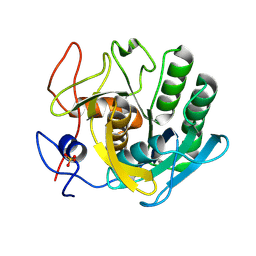 | | PanDDA analysis group deposition -- Proteinase K crystal structure Apo9 | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | Deposit date: | 2020-09-23 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
