5IWW
 
 | | Crystal structure of RNA editing factor of designer PLS-type PPR/9R protein in complex with MORF9/RIP9 | | Descriptor: | Multiple organellar RNA editing factor 9, chloroplastic, PLS9-PPR | | Authors: | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing
Nat Plants, 3, 2017
|
|
7WD2
 
 | | Crystal structure of S43 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Q.H, Gao, G.F, Qi, J.X, Su, C, Liu, H.H, Wu, L.L. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Two pan-SARS-CoV-2 nanobodies and their multivalent derivatives effectively prevent Omicron infections in mice.
Cell Rep Med, 4, 2023
|
|
7WD1
 
 | | Crystal structure of R14 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, R14, Spike protein S1, ... | | Authors: | Wang, Q.H, Gao, G.F, Qi, J.X, Su, C, Liu, H.H, Wu, L.L. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two pan-SARS-CoV-2 nanobodies and their multivalent derivatives effectively prevent Omicron infections in mice.
Cell Rep Med, 4, 2023
|
|
9J52
 
 | | CryoEM structure of human XPR1 in complex with phosphate in state B | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, PHOSPHATE ION, Solute carrier family 53 member 1 | | Authors: | Zhang, W.H, Chen, Y.K, Guan, Z.Y, Liu, Z. | | Deposit date: | 2024-08-11 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the mechanism of phosphate recognition and transport by XPR1.
Nat Commun, 16, 2025
|
|
9J53
 
 | | CryoEM structure of human XPR1 in complex with phosphate in state C | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, PHOSPHATE ION, Solute carrier family 53 member 1 | | Authors: | Zhang, W.H, Chen, Y.K, Guan, Z.Y, Liu, Z. | | Deposit date: | 2024-08-11 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the mechanism of phosphate recognition and transport by XPR1.
Nat Commun, 16, 2025
|
|
9J51
 
 | | CryoEM structure of human XPR1 in complex with phosphate in state A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, PHOSPHATE ION, Solute carrier family 53 member 1 | | Authors: | Zhang, W.H, Chen, Y.K, Guan, Z.Y, Liu, Z. | | Deposit date: | 2024-08-11 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the mechanism of phosphate recognition and transport by XPR1.
Nat Commun, 16, 2025
|
|
4NJL
 
 | | Crystal structure of middle east respiratory syndrome coronavirus S2 protein fusion core | | Descriptor: | S protein, TRIETHYLENE GLYCOL | | Authors: | Zhu, Y, Lu, L, Qin, L, Ye, S, Jiang, S, Zhang, R. | | Deposit date: | 2013-11-10 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor.
Nat Commun, 5, 2014
|
|
7CH5
 
 | |
9INL
 
 | | Crystal structure of SARS-Cov-2 main protease E166R mutant in complex with Bofutrelvir | | Descriptor: | Replicase polyprotein 1a, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhou, X.L, Li, J. | | Deposit date: | 2024-07-08 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|
5IZW
 
 | | Crystal structure of RNA editing specific factor of designer PLS-type PPR-9R protein | | Descriptor: | PLS9-PPR | | Authors: | Yan, J, Zhang, Q, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-26 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing
Nat Plants, 3, 2017
|
|
7CH4
 
 | |
8JIZ
 
 | |
8JJ2
 
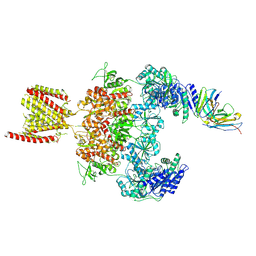 | | Cryo-EM structure of GluN1-2A NMDAR in complex with human Fab2G7 in one fab conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab2G7 Heavy Chain, ... | | Authors: | Wang, H, Zhu, S. | | Deposit date: | 2023-05-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for antibody-mediated NMDA receptor clustering and endocytosis in autoimmune encephalitis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8JJ1
 
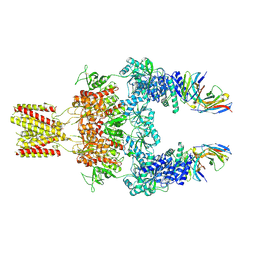 | | Cryo-EM structure of GluN1-2A NMDAR in complex with human Fab2G7 in two fab conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 2G7 Heavy Chain, ... | | Authors: | Wang, H, Zhu, S. | | Deposit date: | 2023-05-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for antibody-mediated NMDA receptor clustering and endocytosis in autoimmune encephalitis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8JJ0
 
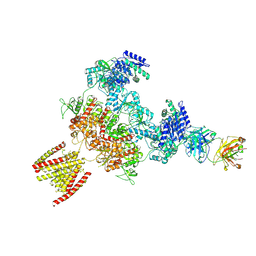 | |
8HDK
 
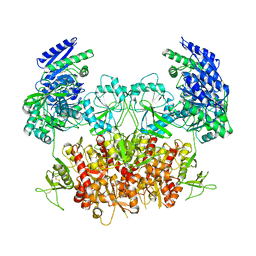 | | Structure of the Rat GluN1-GluN2C NMDA receptor in complex with glycine and glutamate (minor class in symmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-11-04 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8H7Z
 
 | |
8H7L
 
 | | Cryo-EM Structure of SARS-CoV-2 BA.2 Spike protein in complex with BA7535 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7535 fab heavt chain, ... | | Authors: | Liu, Z, Yan, A, Gao, Y. | | Deposit date: | 2022-10-20 | | Release date: | 2023-08-30 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Identification of a highly conserved neutralizing epitope within the RBD region of diverse SARS-CoV-2 variants.
Nat Commun, 15, 2024
|
|
8Y1R
 
 | | in situ room temperature Laue crystallography | | Descriptor: | Lysozyme C | | Authors: | Wang, Z.J, Wang, S.S, Pan, Q.Y, Yu, L, Su, Z.H, Yang, T.Y, Wang, Y.Z, Zhang, W.Z, Hao, Q, Gao, X.Y. | | Deposit date: | 2024-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single pulse data collection with an X-ray chopper at in situ room temperature Laue crystallography beamline BL03HB
Nucl Instrum Methods Phys Res A, 1069, 2024
|
|
7C8J
 
 | | Structural basis for cross-species recognition of COVID-19 virus spike receptor binding domain to bat ACE2 | | Descriptor: | Angiotensin-converting enzyme, SARS-CoV-2 Receptor binding domain, ZINC ION | | Authors: | Liu, K.F, Wang, J, Tan, S.G, Niu, S, Wu, L.L, Zhang, Y.F, Pan, X.Q, Meng, Y.M, Chen, Q, Wang, Q.H, Wang, H.W, Qi, J.X, Gao, G.F. | | Deposit date: | 2020-06-01 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Cross-species recognition of SARS-CoV-2 to bat ACE2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7C8K
 
 | | Structural basis for cross-species recognition of COVID-19 virus spike receptor binding domain to bat ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Liu, K.F, Wang, J, Tan, S.G, Niu, S, Wu, L.L, Zhang, Y.F, Pan, X.Q, Meng, Y.M, Chen, Q, Wang, Q.H, Wang, H.W, Qi, J.X, Gao, G.F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cross-species recognition of SARS-CoV-2 to bat ACE2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6L8O
 
 | | Crystal structure of the K. lactis Rad5 (Hg-derivative) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA repair protein RAD5, MERCURY (II) ION | | Authors: | Shen, M, Xiang, S. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
6L8N
 
 | | Crystal structure of the K. lactis Rad5 | | Descriptor: | DNA repair protein RAD5, ZINC ION | | Authors: | Shen, M, Xiang, S. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
6L8R
 
 | | membrane-bound PD-L1-CD | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Maorong, W, Cao, Y, Bin, W, Bo, O. | | Deposit date: | 2019-11-07 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PD-L1 degradation is regulated by electrostatic membrane association of its cytoplasmic domain.
Nat Commun, 12, 2021
|
|
7YFI
 
 | | Structure of the Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex with glycine and glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
