3LCO
 
 | | Inhibitor Bound to A DFG-Out structure of the Kinase Domain of CSF-1R | | Descriptor: | 3-({4-methoxy-5-[(4-methoxybenzyl)oxy]pyridin-2-yl}methoxy)-5-(1-methyl-1H-pyrazol-4-yl)pyrazin-2-amine, Macrophage colony-stimulating factor 1 receptor | | Authors: | Kamtekar, S, Day, J.E, Reitz, B.A, Mathis, K.J, Meyers, M.J. | | Deposit date: | 2010-01-11 | | Release date: | 2010-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure-based drug design enables conversion of a DFG-in binding CSF-1R kinase inhibitor to a DFG-out binding mode.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3MD9
 
 | | Structure of apo form of a periplasmic heme binding protein | | Descriptor: | BROMIDE ION, GLYCEROL, Hemin-binding periplasmic protein hmuT, ... | | Authors: | Mattle, D, Goetz, B.A, Locher, K.P. | | Deposit date: | 2010-03-30 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Two stacked heme molecules in the binding pocket of the periplasmic heme-binding protein HmuT from Yersinia pestis.
J.Mol.Biol., 404, 2010
|
|
3O00
 
 | | Crystal Structure of the Salmonella Type III Secretion System Tip Protein SipD-C244S | | Descriptor: | Cell invasion protein sipD, NICKEL (II) ION | | Authors: | Chatterjee, S, Zhong, D, Nordhues, B.A, Battaile, K.P, Lovell, S, DeGuzman, R.N. | | Deposit date: | 2010-07-18 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate.
Protein Sci., 20, 2011
|
|
3O2U
 
 | | S. cerevisiae Ubc12 | | Descriptor: | GLYCEROL, NEDD8-conjugating enzyme UBC12 | | Authors: | Scott, D.C, Monda, J.K, Grace, C.R.R, Duda, D.M, Kriwacki, R.W, Kurz, T, Schulman, B.A. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | A dual E3 mechanism for Rub1 ligation to Cdc53.
Mol.Cell, 39, 2010
|
|
3O2P
 
 | | A Dual E3 Mechanism for Rub1 Ligation to Cdc53: Dcn1(P)-Cdc53(WHB) | | Descriptor: | Cell division control protein 53, Defective in cullin neddylation protein 1, GLYCEROL | | Authors: | Scott, D.C, Monda, J.K, Grace, C.R.R, Duda, D.M, Kriwacki, R.W, Kurz, T, Schulman, B.A. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | A dual E3 mechanism for Rub1 ligation to Cdc53.
Mol.Cell, 39, 2010
|
|
3MID
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound azide obtained by soaking (100mM NaN3) | | Descriptor: | AZIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
3MIH
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound azide, obtained in the presence of substrate | | Descriptor: | AZIDE ION, COPPER (II) ION, IODIDE ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
3MLK
 
 | | Reduced (Cu+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound nitrite | | Descriptor: | COPPER (II) ION, NICKEL (II) ION, NITRITE ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-16 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Differential reactivity between two copper sites in peptidylglycine alpha-hydroxylating monooxygenase
J.Am.Chem.Soc., 132, 2010
|
|
3MIE
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound azide obtained by soaking (50mM NaN3) | | Descriptor: | AZIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
3MLJ
 
 | | Reduced (Cu+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound carbon monooxide (CO) | | Descriptor: | ACETATE ION, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Prigge, S.T, Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-16 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Differential reactivity between two copper sites in peptidylglycine alpha-hydroxylating monooxygenase
J.Am.Chem.Soc., 132, 2010
|
|
3MIG
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound nitrite, obtained in the presence of substrate | | Descriptor: | COPPER (II) ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
3ME0
 
 | | Structure of the E. coli chaperone PAPD in complex with the pilin domain of the PapGII adhesin | | Descriptor: | Chaperone protein papD, PapG protein | | Authors: | Ford, B.A, Elam, J.S, Dodson, K.W, Pinkner, J.S, Hultgren, S.J. | | Deposit date: | 2010-03-31 | | Release date: | 2011-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of the E. Coli Chaperone PapD in Complex with the Pilin Domain of the PapGII Adhesin
To be Published
|
|
3NU1
 
 | | Structure of holo form of a periplasmic heme binding protein | | Descriptor: | Hemin-binding periplasmic protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mattle, D, Goetz, B.A, Woo, J.S, Locher, K.P. | | Deposit date: | 2010-07-06 | | Release date: | 2010-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two stacked heme molecules in the binding pocket of the periplasmic heme-binding protein HmuT from Yersinia pestis.
J.Mol.Biol., 404, 2010
|
|
3O02
 
 | | The Crystal Structure of the Salmonella Type III Secretion System Tip Protein SipD in Complex with Chenodeoxycholate | | Descriptor: | CHENODEOXYCHOLIC ACID, Cell invasion protein sipD, NICKEL (II) ION | | Authors: | Chatterjee, S, Zhong, D, Nordhues, B.A, Battaile, K.P, Lovell, S, DeGuzman, R.N. | | Deposit date: | 2010-07-18 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate.
Protein Sci., 20, 2011
|
|
3O01
 
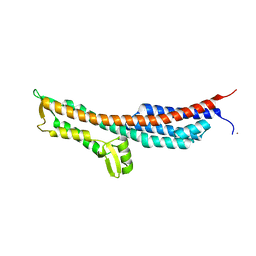 | | The Crystal Structure of the Salmonella Type III Secretion System Tip Protein SipD in Complex with Deoxycholate | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, Cell invasion protein sipD, NICKEL (II) ION | | Authors: | Chatterjee, S, Zhong, D, Nordhues, B.A, Battaile, K.P, Lovell, S, DeGuzman, R.N. | | Deposit date: | 2010-07-18 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate.
Protein Sci., 20, 2011
|
|
3MLL
 
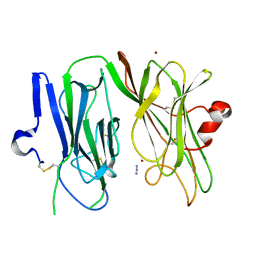 | | Reduced (Cu+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound azide | | Descriptor: | AZIDE ION, COPPER (II) ION, NICKEL (II) ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-16 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Differential reactivity between two copper sites in peptidylglycine alpha-hydroxylating monooxygenase
J.Am.Chem.Soc., 132, 2010
|
|
3MMU
 
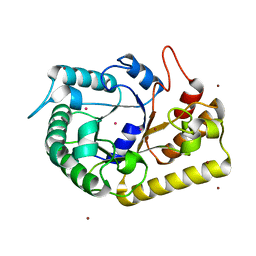 | | Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima | | Descriptor: | CADMIUM ION, Endoglucanase, NICKEL (II) ION | | Authors: | Pereira, J.H, Chen, Z, McAndrew, R.P, Sapra, R, Chhabra, S.R, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2010-04-20 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Biochemical characterization and crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima.
J.Struct.Biol., 172, 2010
|
|
3MIB
 
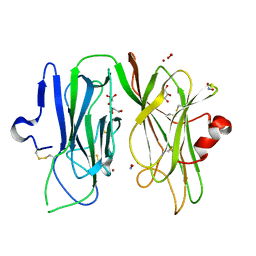 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound nitrite | | Descriptor: | COPPER (II) ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
3N4D
 
 | |
3OIL
 
 | | Human Carbonic anhydrase II mutant A65S, N67Q (CA IX mimic) bound by 2-Ethylestradiol 3-O-sulfamate | | Descriptor: | (14beta,17alpha)-2-ethyl-17-hydroxyestra-1(10),2,4-trien-3-yl sulfamate, Carbonic anhydrase 2, ZINC ION | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
3O6B
 
 | | A Dual E3 Mechanism for Rub1 Ligation to Cdc53: Dcn1(P)-Cdc53(WHB) low resolution | | Descriptor: | Cell division control protein 53, Defective in cullin neddylation protein 1 | | Authors: | Scott, D.C, Monda, J.K, Grace, C.R.R, Duda, D.M, Kriwacki, R.W, Kurz, T, Schulman, B.A. | | Deposit date: | 2010-07-28 | | Release date: | 2010-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A dual E3 mechanism for Rub1 ligation to Cdc53.
Mol.Cell, 39, 2010
|
|
3NZZ
 
 | | Crystal Structure of the Salmonella Type III Secretion System Tip Protein SipD | | Descriptor: | Cell invasion protein sipD, NICKEL (II) ION | | Authors: | Chatterjee, S, Zhong, D, Nordhues, B.A, Battaile, K.P, Lovell, S, DeGuzman, R.N. | | Deposit date: | 2010-07-18 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate.
Protein Sci., 20, 2011
|
|
3OKU
 
 | | Human Carbonic Anhydrase II in complex with 2-Ethylestrone-3-O-sulfamate | | Descriptor: | (9beta)-2-ethyl-17-oxoestra-1(10),2,4-trien-3-yl sulfamate, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-25 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
3OIM
 
 | | Human Carbonic anhydrase II bound by 2-Ethylestradiol 3-O-sulfamate | | Descriptor: | (14beta,17alpha)-2-ethyl-17-hydroxyestra-1(10),2,4-trien-3-yl sulfamate, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
3PAR
 
 | | Surfactant Protein-A neck and carbohydrate recognition domain (NCRD) in the absence of ligand | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein A, SULFATE ION | | Authors: | Shang, F, Rynkiewicz, M.J, McCormack, F.X, Wu, H, Cafarella, T.M, Head, J, Seaton, B.A. | | Deposit date: | 2010-10-19 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic complexes of surfactant protein A and carbohydrates reveal ligand-induced conformational change.
J.Biol.Chem., 286, 2011
|
|
