3AZF
 
 | | Crystal Structure of Human Nucleosome Core Particle Containing H3K79Q mutation | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2011-05-25 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
3AZE
 
 | | Crystal Structure of Human Nucleosome Core Particle Containing H3K64Q mutation | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2011-05-25 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
3AZG
 
 | | Crystal Structure of Human Nucleosome Core Particle Containing H3K115Q mutation | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2011-05-25 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
3WFI
 
 | | The crystal structure of D-mandelate dehydrogenase | | Descriptor: | 2-dehydropantoate 2-reductase | | Authors: | Miyanaga, A, Fujisawa, S, Furukawa, N, Arai, K, Nakajima, M, Taguchi, H. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | The crystal structure of D-mandelate dehydrogenase reveals its distinct substrate and coenzyme recognition mechanisms from those of 2-ketopantoate reductase.
Biochem.Biophys.Res.Commun., 439, 2013
|
|
3AZK
 
 | | Crystal Structure of Human Nucleosome Core Particle Containing H4K59Q mutation | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2011-05-25 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
3WMU
 
 | | The structure of an anti-cancer lectin mytilec apo-form from the mussel Mytilus galloprovincialis | | Descriptor: | GLYCEROL, Lectin | | Authors: | Terada, D, Kawai, F, Noguchi, H, Unzai, S, Park, S.-Y, Ozeki, Y, Tame, J.R.H. | | Deposit date: | 2013-11-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of MytiLec, a galactose-binding lectin from the mussel Mytilus galloprovincialis with cytotoxicity against certain cancer cell types
Sci Rep, 6, 2016
|
|
3AZJ
 
 | | Crystal Structure of Human Nucleosome Core Particle Containing H4K44Q mutation | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2011-05-25 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
3WVL
 
 | | Crystal structure of the football-shaped GroEL-GroES complex (GroEL: GroES2:ATP14) from Escherichia coli | | Descriptor: | 10 kDa chaperonin, 60 kDa chaperonin, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Koike-Takeshita, A, Arakawa, T, Taguchi, H, Shimamura, T. | | Deposit date: | 2014-05-23 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.788 Å) | | Cite: | Crystal structure of a symmetric football-shaped GroEL:GroES2-ATP14 complex determined at 3.8 angstrom reveals rearrangement between two GroEL rings.
J.Mol.Biol., 426, 2014
|
|
3WW8
 
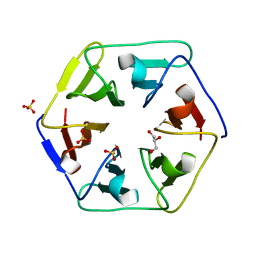 | | Crystal structure of the computationally designed Pizza3 protein | | Descriptor: | GLYCEROL, Pizza3 protein, SULFATE ION | | Authors: | Voet, A.R.D, Noguchi, H, Addy, C, Simoncini, D, Terada, D, Unzai, S, Park, S.Y, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2014-06-17 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Computational design of a self-assembling symmetrical beta-propeller protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2ZBO
 
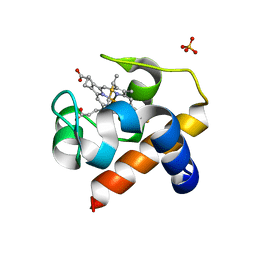 | | Crystal structure of low-redox-potential cytochrom c6 from brown alga Hizikia fusiformis at 1.6 A resolution | | Descriptor: | Cytochrome c6, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Akazaki, H, Kawai, F, Chida, H, Matsumoto, Y, Sirasaki, I, Nakade, H, Hirayama, M, Hosikawa, K, Suruga, K, Satoh, T, Yamada, S, Unzai, S, Hakamata, W, Nishio, T, Park, S.-Y, Oku, T. | | Deposit date: | 2007-10-26 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cloning, expression and purification of cytochrome c(6) from the brown alga Hizikia fusiformis and complete X-ray diffraction analysis of the structure
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
2ZNP
 
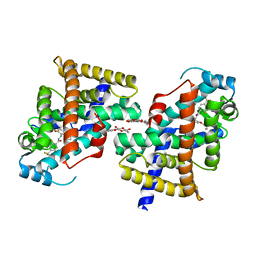 | | Human PPAR delta ligand binding domain in complex with a synthetic agonist TIPP204 | | Descriptor: | (2S)-2-{4-butoxy-3-[({[2-fluoro-4-(trifluoromethyl)phenyl]carbonyl}amino)methyl]benzyl}butanoic acid, Peroxisome proliferator-activated receptor delta, heptyl beta-D-glucopyranoside | | Authors: | Oyama, T, Hirakawa, Y, Nagasawa, N, Miyachi, H, Morikawa, K. | | Deposit date: | 2008-04-30 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Adaptability and selectivity of human peroxisome proliferator-activated receptor (PPAR) pan agonists revealed from crystal structures
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3A4S
 
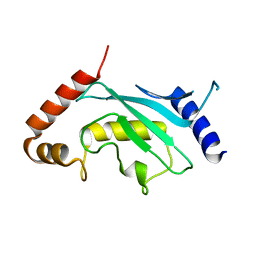 | | The crystal structure of the SLD2:Ubc9 complex | | Descriptor: | NFATC2-interacting protein, SUMO-conjugating enzyme UBC9 | | Authors: | Sekiyama, N, Arita, K, Ikeda, Y, Ariyoshi, M, Tochio, H, Saitoh, H, Shirakawa, M. | | Deposit date: | 2009-07-14 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for regulation of poly-SUMO chain by a SUMO-like domain of Nip45
Proteins, 78, 2009
|
|
2DKH
 
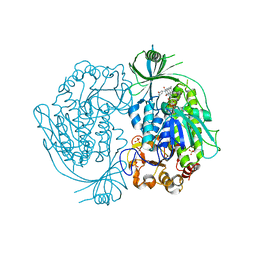 | | Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni, in complex with the substrate | | Descriptor: | 3-HYDROXYBENZOIC ACID, 3-hydroxybenzoate hydroxylase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hiromoto, T, Fujiwara, S, Hosokawa, K, Yamaguchi, H. | | Deposit date: | 2006-04-11 | | Release date: | 2006-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni has a large tunnel for substrate and oxygen access to the active site
J.Mol.Biol., 364, 2006
|
|
3AKG
 
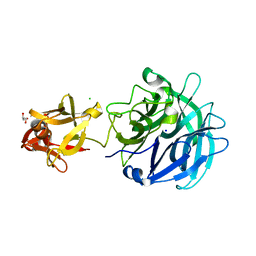 | | Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-1,5-L-arabinofuranobiose | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative secreted alpha L-arabinofuranosidase II, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of an Exo-1,5-{alpha}-L-arabinofuranosidase from Streptomyces avermitilis Provides Insights into the Mechanism of Substrate Discrimination between Exo- and Endo-type Enzymes in Glycoside Hydrolase Family 43.
J.Biol.Chem., 285, 2010
|
|
2ZNQ
 
 | | Human PPAR delta ligand binding domain in complex with a synthetic agonist TIPP401 | | Descriptor: | (2S)-2-{3-[({[2-fluoro-4-(trifluoromethyl)phenyl]carbonyl}amino)methyl]-4-methoxybenzyl}butanoic acid, Peroxisome proliferator-activated receptor delta, heptyl beta-D-glucopyranoside | | Authors: | Oyama, T, Hirakawa, Y, Nagasawa, N, Miyachi, H, Morikawa, K. | | Deposit date: | 2008-04-30 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Adaptability and selectivity of human peroxisome proliferator-activated receptor (PPAR) pan agonists revealed from crystal structures
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3AKH
 
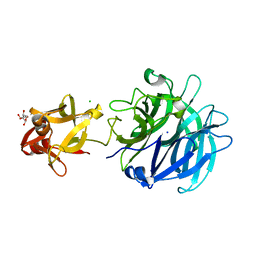 | | Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-1,5-L-arabinofuranotriose | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative secreted alpha L-arabinofuranosidase II, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of an Exo-1,5-{alpha}-L-arabinofuranosidase from Streptomyces avermitilis Provides Insights into the Mechanism of Substrate Discrimination between Exo- and Endo-type Enzymes in Glycoside Hydrolase Family 43.
J.Biol.Chem., 285, 2010
|
|
3AKF
 
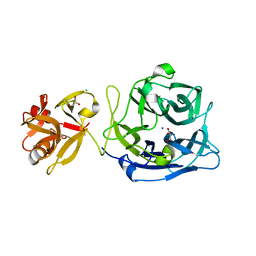 | | Crystal structure of exo-1,5-alpha-L-arabinofuranosidase | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative secreted alpha L-arabinofuranosidase II, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of an Exo-1,5-{alpha}-L-arabinofuranosidase from Streptomyces avermitilis Provides Insights into the Mechanism of Substrate Discrimination between Exo- and Endo-type Enzymes in Glycoside Hydrolase Family 43.
J.Biol.Chem., 285, 2010
|
|
2Z5V
 
 | | Solution structure of the TIR domain of human MyD88 | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Ohnishi, H, Tochio, H, Hiroaki, H, Kondo, N, Kato, Z, Shirakawa, M. | | Deposit date: | 2007-07-19 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the multiple interactions of the MyD88 TIR domain in TLR4 signaling.
Proc.Natl.Acad.Sci.USA, 2009
|
|
2ZNN
 
 | | Human PPAR alpha ligand binding domain in complex with a synthetic agonist TIPP703 | | Descriptor: | (2S)-2-(4-propoxy-3-{[({4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]phenyl}carbonyl)amino]methyl}benzyl)butanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Oyama, T, Toyota, K, Kasuga, J, Miyachi, H, Morikawa, K. | | Deposit date: | 2008-04-30 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Adaptability and selectivity of human peroxisome proliferator-activated receptor (PPAR) pan agonists revealed from crystal structures
Acta Crystallogr.,Sect.D, 65, 2009
|
|
7DC8
 
 | | Crystal structure of Switch Ab Fab and hIL6R in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Interleukin-6 receptor subunit alpha, SULFATE ION, ... | | Authors: | Kadono, S, Fukami, T.A, Kawauchi, H, Torizawa, T, Mimoto, F. | | Deposit date: | 2020-10-23 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.757 Å) | | Cite: | Exploitation of Elevated Extracellular ATP to Specifically Direct Antibody to Tumor Microenvironment.
Cell Rep, 33, 2020
|
|
2D1O
 
 | | Stromelysin-1 (MMP-3) complexed to a hydroxamic acid inhibitor | | Descriptor: | CALCIUM ION, SM-25453, Stromelysin-1, ... | | Authors: | Kohno, T, Hochigai, H, Yamashita, E, Tsukihara, T, Kanaoka, M. | | Deposit date: | 2005-08-30 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structures of the catalytic domain of human stromelysin-1 (MMP-3) and collagenase-3 (MMP-13) with a hydroxamic acid inhibitor SM-25453
Biochem.Biophys.Res.Commun., 344, 2006
|
|
2D04
 
 | | Crystal structure of neoculin, a sweet protein with taste-modifying activity. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Curculin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shimizu-Ibuka, A, Morita, Y, Terada, T, Asakura, T, Nakajima, K, Iwata, S, Misaka, T, Sorimachi, H, Arai, S, Abe, K. | | Deposit date: | 2005-07-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Crystal structure of neoculin: insights into its sweetness and taste-modifying activity
J.Mol.Biol., 359, 2006
|
|
2DKI
 
 | | Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni, under pressure of xenon gas (12 atm) | | Descriptor: | 3-HYDROXYBENZOATE HYDROXYLASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Hiromoto, T, Fujiwara, S, Hosokawa, K, Yamaguchi, H. | | Deposit date: | 2006-04-11 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni has a large tunnel for substrate and oxygen access to the active site
J.Mol.Biol., 364, 2006
|
|
3AKI
 
 | | Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-L-arabinofuranosyl azido | | Descriptor: | (2R,3R,4R,5S)-2-azido-5-(hydroxymethyl)oxolane-3,4-diol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of an Exo-1,5-{alpha}-L-arabinofuranosidase from Streptomyces avermitilis Provides Insights into the Mechanism of Substrate Discrimination between Exo- and Endo-type Enzymes in Glycoside Hydrolase Family 43.
J.Biol.Chem., 285, 2010
|
|
2DFK
 
 | | Crystal structure of the CDC42-Collybistin II complex | | Descriptor: | GLYCEROL, SULFATE ION, cell division cycle 42 isoform 1, ... | | Authors: | Xiang, S, Kim, E.Y, Connelly, J.J, Nassar, N, Kirsch, J, Winking, J, Schwarz, G, Schindelin, H. | | Deposit date: | 2006-03-02 | | Release date: | 2006-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of Cdc42 in Complex with Collybistin II, a Gephyrin-interacting Guanine Nucleotide Exchange Factor.
J.Mol.Biol., 359, 2006
|
|
