5IR2
 
 | | Crystal structure of novel cellulases from microbes associated with the gut ecosystem | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Cellulase, ... | | Authors: | Chang, C, Mack, J, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-03-11 | | Release date: | 2016-03-23 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Crystal structure of novel cellulases from microbes associated with the gut ecosystem
To Be Published
|
|
5I4Q
 
 | | Contact-dependent inhibition system from Escherichia coli NC101 - ternary CdiA/CdiI/EF-Tu complex (domains 2 and 3) | | Descriptor: | CHLORIDE ION, Contact-dependent inhibitor A, Contact-dependent inhibitor I, ... | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2016-02-12 | | Release date: | 2017-06-28 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a novel antibacterial toxin that exploits elongation factor Tu to cleave specific transfer RNAs.
Nucleic Acids Res., 45, 2017
|
|
7STS
 
 | | Crystal Structure of Human Fab S24-1379 in the Complex with the N-teminal Domain of Nucleocapsid Protein from SARS CoV-2 | | Descriptor: | Fab S24-1379, heavy chain, light chain, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7SUE
 
 | | Crystal Structure of Human Fab S24-188 in the complex with the N-teminal Domain of Nucleocapsid protein from SARS CoV-2 | | Descriptor: | Nucleoprotein, S24-188 Fab Heavy chain, S24-188 Fab Light chain | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-17 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
6NAU
 
 | | 1.55 Angstrom Resolution Crystal Structure of 6-phosphogluconolactonase from Klebsiella pneumoniae | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-phosphogluconolactonase, CHLORIDE ION | | Authors: | Minasov, G, Shuvalova, L, Pshenychnyi, S, Dubrovska, I, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-06 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
7STR
 
 | | Crystal Structure of Human Fab S24-1063 in the Complex with the N-teminal Domain of Nucleocapsid Protein from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Fab S24-1063, Heavy chain, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
6NBG
 
 | | 2.05 Angstrom Resolution Crystal Structure of Hypothetical Protein KP1_5497 from Klebsiella pneumoniae. | | Descriptor: | CHLORIDE ION, Glucosamine-6-phosphate deaminase, PHOSPHATE ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6WGQ
 
 | | The crystal structure of a beta-lactamase from Shigella flexneri 2a str. 2457T | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-06 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of a beta-lactamase from Shigella flexneri 2a str. 2457T
To Be Published
|
|
1KTN
 
 | | Structural Genomics, Protein EC1535 | | Descriptor: | 2-deoxyribose-5-phosphate aldolase | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Skarina, T, Evdokimova, E, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-16 | | Release date: | 2002-08-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.5A crystal structure of
2-deoxyribose-5-phosphate aldlase
To be Published
|
|
6W4B
 
 | | The crystal structure of Nsp9 RNA binding protein of SARS CoV-2 | | Descriptor: | Non-structural protein 9 | | Authors: | Tan, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-10 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of Nsp9 replicase protein of COVID-19
To Be Published
|
|
6OYF
 
 | | The structure of condensation and adenylation domains of teixobactin-producing nonribosomal peptide synthetase Txo1 serine module | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMIC ACID, ... | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-14 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of teixobactin-producing nonribosomal peptide synthetase condensation and adenylation domains.
Curr Res Struct Biol, 2, 2020
|
|
5HG0
 
 | | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis complex with SAM | | Descriptor: | Pantothenate synthetase, S-ADENOSYLMETHIONINE | | Authors: | Chang, C, Maltseva, N, Kim, Y, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-07 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis complex with SAM
To Be Published
|
|
6WZU
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , P3221 space group | | Descriptor: | CHLORIDE ION, GLYCEROL, Non-structural protein 3, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-14 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
5HI6
 
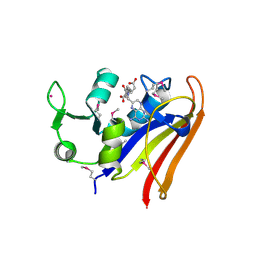 | | The high resolution structure of dihydrofolate reductase from Yersinia pestis complex with methotrexate as closed form | | Descriptor: | CALCIUM ION, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-11 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | The high resolution structure of dihydrofolate reductase from Yersinia pestis complex with methotrexate as closed form
To Be Published
|
|
6N7M
 
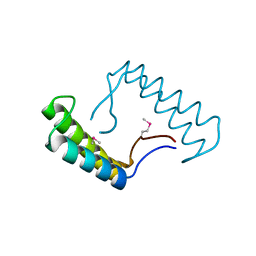 | | 1.78 Angstrom Resolution Crystal Structure of Hypothetical Protein CD630_05490 from Clostridioides difficile 630. | | Descriptor: | Hypothetical Protein CD630_05490 | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Dubrovska, I, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-27 | | Release date: | 2018-12-12 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 1.78 Angstrom Resolution Crystal Structure of Hypothetical Protein CD630_05490 from Clostridioides difficile 630.
To Be Published
|
|
5HD6
 
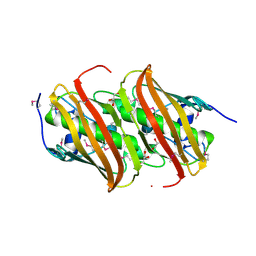 | | High resolution structure of 3-hydroxydecanoyl-(acyl carrier protein) dehydratase from Yersinia pestis at 1.35 A | | Descriptor: | 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase, GLYCEROL | | Authors: | Chang, C, Maltseva, N, Kim, Y, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-04 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High resolution structure of 3-hydroxydecanoyl-(acyl carrier protein) dehydratase from Yersinia pestis at 1.35 A
To Be Published
|
|
6WT2
 
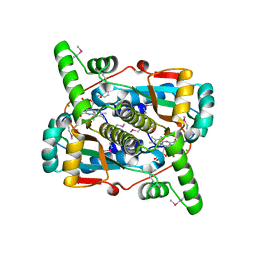 | | Crystal Structure of Putative NAD(P)H-Flavin Oxidoreductase from Neisseria meningitidis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Crofts, T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-01 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional and Structural Characterization of Diverse NfsB Chloramphenicol Reductase Enzymes from Human Pathogens.
Microbiol Spectr, 10, 2022
|
|
6WHL
 
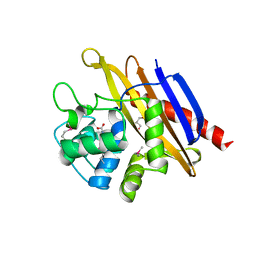 | |
6WEN
 
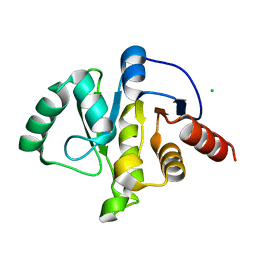 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in the apo form | | Descriptor: | CHLORIDE ION, Non-structural protein 3 | | Authors: | Michalska, K, Stols, L, Jedrzejczak, R, Endres, M, Babnigg, G, Kim, Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-02 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
6WGR
 
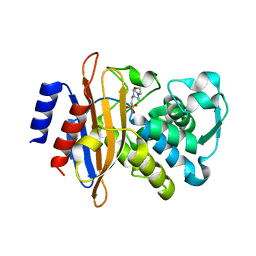 | | The crystal structure of a beta-lactamase from Staphylococcus aureus subsp. aureus USA300_TCH1516 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase, GLYCEROL | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-06 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The crystal structure of a beta-lactamase from Staphylococcus aureus subsp. aureus USA300_TCH1516
To Be Published
|
|
5FDA
 
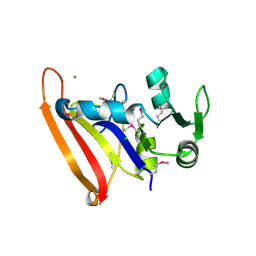 | | The high resolution structure of apo form dihydrofolate reductase from Yersinia pestis at 1.55 A | | Descriptor: | CHLORIDE ION, Dihydrofolate reductase | | Authors: | Chang, C, Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-15 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | structure of dihydrofolate reductase from Yersinia pestis complex with
To Be Published
|
|
3K0L
 
 | | Crystal Structure of Putative MarR Family Transcriptional Regulator from Acinetobacter sp. ADP | | Descriptor: | Repressor protein, SULFATE ION | | Authors: | Kim, Y, Joachimiak, G, Bigelow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-24 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Crystal Structure of Putative MarR Family Transcriptional Regulator from Acinetobacter sp. ADP
To be Published
|
|
1R4V
 
 | | 1.9A crystal structure of protein AQ328 from Aquifex aeolicus | | Descriptor: | CACODYLATE ION, Hypothetical protein AQ_328, ZINC ION | | Authors: | Qiu, Y, Tereshko, V, Kim, Y, Zhang, R, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-10-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of Aq_328 from the hyperthermophilic bacteria Aquifex aeolicus shows an ancestral histone fold.
Proteins, 62, 2006
|
|
1UXO
 
 | | The crystal structure of the ydeN gene product from B. subtilis | | Descriptor: | Putative hydrolase YdeN | | Authors: | Janda, I.K, Devedjiev, Y, Cooper, D.R, Chruszcz, M, Derewenda, U, Gabrys, A, Minor, W, Joachimiak, A, Derewenda, Z.S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-27 | | Release date: | 2004-05-27 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Harvesting the high-hanging fruit: the structure of the YdeN gene product from Bacillus subtilis at 1.8 angstroms resolution.
Acta Crystallogr. D Biol. Crystallogr., 60, 2004
|
|
5I4R
 
 | | Contact-dependent inhibition system from Escherichia coli NC101 - ternary CdiA/CdiI/EF-Tu complex (trypsin-modified) | | Descriptor: | Contact-dependent inhibitor A, Contact-dependent inhibitor I, Elongation factor Tu, ... | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2016-02-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of a novel antibacterial toxin that exploits elongation factor Tu to cleave specific transfer RNAs.
Nucleic Acids Res., 45, 2017
|
|
