6EUZ
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (MK37) | | Descriptor: | (3~{R})-8-methoxy-7-(naphthalen-1-ylmethyl)-5-oxidanylidene-2,3-dihydro-[1,3]thiazolo[3,2-a]pyridine-3-carboxylic acid, ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, ... | | Authors: | Begum, A, Hall, M, Grundstrom, C, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-10-31 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
6EXL
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (MK206) - folded HTH motif | | Descriptor: | ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, SODIUM ION, ... | | Authors: | Hall, M, Grundstrom, C, Begum, A, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-11-08 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
6EUU
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (KSK29) | | Descriptor: | (3~{R})-8-cyclopropyl-5-oxidanylidene-7-(quinolin-3-ylmethyl)-2,3-dihydro-[1,3]thiazolo[3,2-a]pyridine-3-carboxylic acid, ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, ... | | Authors: | Begum, A, Hall, M, Grundstrom, C, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-10-31 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
6EV0
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (AC129) | | Descriptor: | ISOPROPYL ALCOHOL, Listeriolysin regulatory protein, [(3~{R})-3-carboxy-7-(naphthalen-1-ylmethyl)-5-oxidanylidene-2,3-dihydro-[1,3]thiazolo[3,2-a]pyridin-8-yl]-dimethyl-azanium, ... | | Authors: | Begum, A, Hall, M, Grundstrom, C, Cairns, A.G, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-10-31 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
6EYX
 
 | |
6EY4
 
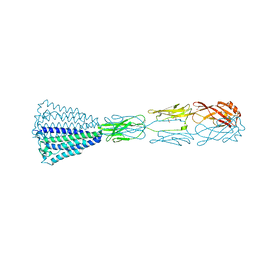 | | Periplasmic domain (residues 36-513) of GldM | | Descriptor: | 1,2-ETHANEDIOL, GldM | | Authors: | Leone, P, Roche, J, Cambillau, C, Roussel, A. | | Deposit date: | 2017-11-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Type IX secretion system PorM and gliding machinery GldM form arches spanning the periplasmic space.
Nat Commun, 9, 2018
|
|
6EU7
 
 | |
6EXK
 
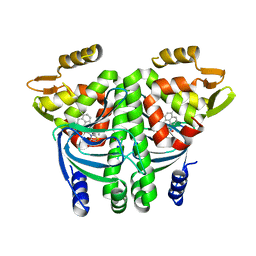 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (MK206) - unfolded HTH motif | | Descriptor: | ISOPROPYL ALCOHOL, Listeriolysin regulatory protein, SODIUM ION, ... | | Authors: | Hall, M, Grundstrom, C, Begum, A, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-11-08 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
6FLG
 
 | |
6EZ9
 
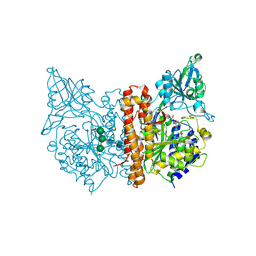 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) - the E424M inactive mutant, in complex with a inhibitor JHU3372 | | Descriptor: | (2~{S})-2-[[(2~{S})-4-methyl-1-oxidanyl-1-oxidanylidene-pentan-2-yl]oxycarbonylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C, Novakova, Z, Motlova, L. | | Deposit date: | 2017-11-14 | | Release date: | 2018-12-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural and computational basis for potent inhibition of glutamate carboxypeptidase II by carbamate-based inhibitors.
Bioorg.Med.Chem., 27, 2019
|
|
4LBH
 
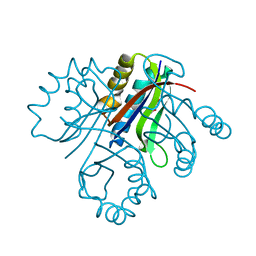 | | 5-chloro-2-hydroxyhydroquinone dehydrochlorinase (TftG) from Burkholderia phenoliruptrix AC1100: Apo-form | | Descriptor: | 5-chloro-2-hydroxyhydroquinone dehydrochlorinase (TftG), DI(HYDROXYETHYL)ETHER | | Authors: | Hayes, R.P, Lewis, K.M, Xun, L, Kang, C. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Catalytic Mechanism of 5-Chlorohydroxyhydroquinone Dehydrochlorinase from the YCII Superfamily of Largely Unknown Function.
J.Biol.Chem., 288, 2013
|
|
8ALK
 
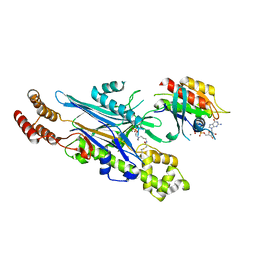 | | Structure of the Legionella phosphocholine hydrolase Lem3 in complex with its substrate Rab1 | | Descriptor: | 2-[[[5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxyethyl-[3-(2-chloranylethanoylamino)propyl]-dimethyl-azanium, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kaspers, M.S, Pett, C, Hedberg, C, Itzen, A, Pogenberg, V. | | Deposit date: | 2022-08-01 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dephosphocholination by Legionella effector Lem3 functions through remodelling of the switch II region of Rab1b.
Nat Commun, 14, 2023
|
|
8ANP
 
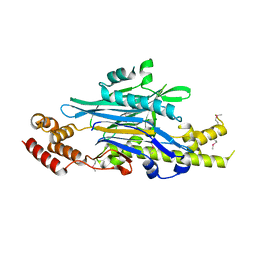 | | Legionella effector Lem3 mutant D190A in complex with Mg2+ | | Descriptor: | MAGNESIUM ION, Phosphocholine hydrolase Lem3, SULFATE ION, ... | | Authors: | Kaspers, M.S, Pogenberg, V, Ernst, S, Ecker, F, Pett, C, Ochtrop, P, Hedberg, C, Groll, M, Itzen, A. | | Deposit date: | 2022-08-05 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dephosphocholination by Legionella effector Lem3 functions through remodelling of the switch II region of Rab1b.
Nat Commun, 14, 2023
|
|
1KHF
 
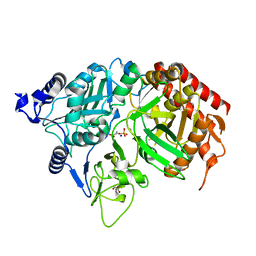 | | PEPCK complex with PEP | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Dunten, P, Belunis, C, Crowther, R, Hollfelder, K, Kammlott, U, Levin, W, Michel, H, Ramsey, G.B, Swain, A, Weber, D, Wertheimer, S.J. | | Deposit date: | 2001-11-29 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of human cytosolic phosphoenolpyruvate carboxykinase reveals a new GTP-binding site.
J.Mol.Biol., 316, 2002
|
|
1KQE
 
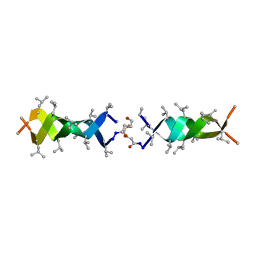 | | Solution structure of a linked shortened gramicidin A in benzene/acetone 10:1 | | Descriptor: | MINI-GRAMICIDIN A | | Authors: | Arndt, H.D, Bockelmann, D, Knoll, A, Lamberth, S, Griesinger, C, Koert, U. | | Deposit date: | 2002-01-05 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Cation Control in Functional Helical Programming: Structures of a D,L-Peptide Ion Channel
Angew.Chem.Int.Ed.Engl., 41, 2002
|
|
2I9S
 
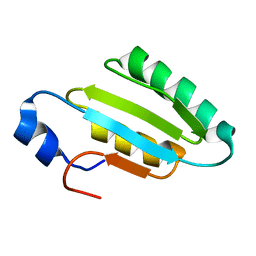 | | The solution structure of the core of mesoderm development (MESD). | | Descriptor: | Mesoderm development candidate 2 | | Authors: | Koehler, C, Andersen, O, Diehl, A, Schmieder, P, Krause, G, Oschkinat, H. | | Deposit date: | 2006-09-06 | | Release date: | 2007-05-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the core of mesoderm development (MESD), a chaperone for members of the LDLR-family
J.STRUCT.FUNCT.GENOM., 7, 2006
|
|
4LKQ
 
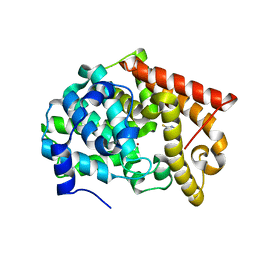 | | Crystal structure of PDE10A2 with fragment ZT017 | | Descriptor: | 4-amino-1,7-dihydro-6H-pyrazolo[3,4-d]pyrimidine-6-thione, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-08 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
1KVN
 
 | | Solution Structure Of Protein SRP19 Of The Arhaeoglobus fulgidus Signal Recognition Particle, 10 Structures | | Descriptor: | SRP19 | | Authors: | Pakhomova, O.N, Deep, S, Huang, Q, Zwieb, C, Hinck, A.P. | | Deposit date: | 2002-01-27 | | Release date: | 2002-03-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein SRP19 of Archaeoglobus fulgidus signal recognition particle.
J.Mol.Biol., 317, 2002
|
|
4LMM
 
 | | Crystal structure of NHERF1 PDZ1 domain complexed with the CXCR2 C-terminal tail in P21 space group | | Descriptor: | ACETIC ACID, CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Jiang, Y, Lu, G, Wu, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-07-10 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
1KXV
 
 | | Camelid VHH Domains in Complex with Porcine Pancreatic alpha-Amylase | | Descriptor: | ALPHA-AMYLASE, PANCREATIC, CAMELID VHH DOMAIN CAB10 | | Authors: | Desmyter, A, Spinelli, S, Payan, F, Lauwereys, M, Wyns, L, Muyldermans, S, Cambillau, C. | | Deposit date: | 2002-02-01 | | Release date: | 2002-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology.
J.Biol.Chem., 277, 2002
|
|
1KXG
 
 | | The 2.0 Ang Resolution Structure of BLyS, B Lymphocyte Stimulator. | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, B lymphocyte stimulator, CITRIC ACID, ... | | Authors: | Oren, D.A, Li, Y, Volovik, Y, Morris, T.S, Dharia, C, Das, K, Galperina, O, Gentz, R, Arnold, E. | | Deposit date: | 2002-01-31 | | Release date: | 2002-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of BLyS receptor recognition.
Nat.Struct.Biol., 9, 2002
|
|
4LBI
 
 | |
1KFL
 
 | | Crystal structure of phenylalanine-regulated 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHP synthase) from E.coli complexed with Mn2+, PEP, and Phe | | Descriptor: | 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase, MANGANESE (II) ION, PHENYLALANINE, ... | | Authors: | Shumilin, I.A, Zhao, C, Bauerle, R, Kretsinger, R.H. | | Deposit date: | 2001-11-21 | | Release date: | 2002-08-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric inhibition of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase alters the coordination of both substrates.
J.Mol.Biol., 320, 2002
|
|
2I7U
 
 | | Structural and Dynamical Analysis of a Four-Alpha-Helix Bundle with Designed Anesthetic Binding Pockets | | Descriptor: | Four-alpha-helix bundle | | Authors: | Ma, D, Brandon, N.R, Cui, T, Bondarenko, V, Canlas, C, Johansson, J.S, Tang, P, Xu, Y. | | Deposit date: | 2006-08-31 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Four-alpha-helix bundle with designed anesthetic binding pockets. Part I: structural and dynamical analyses.
Biophys.J., 94, 2008
|
|
4LDG
 
 | | Crystal Structure of CpSET8 from Cryptosporidium, cgd4_370 | | Descriptor: | MALONATE ION, Protein with a SET domain within carboxy region | | Authors: | Wernimont, A.K, Tempel, W, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, El Bakkouri, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of CpSET8 from Cryptosporidium, cgd4_370
To be Published
|
|
