5A9X
 
 | | Structure of GDP bound BipA | | Descriptor: | GTP-BINDING PROTEIN, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8E2B
 
 | | N-terminal domain of S. aureus GpsB | | Descriptor: | Cell cycle protein GpsB, GLYCEROL | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2022-08-14 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Staphylococcus aureus FtsZ and PBP4 bind to the conformationally dynamic N-terminal domain of GpsB.
Elife, 13, 2024
|
|
8E2C
 
 | |
5AA0
 
 | | Complex of Thermous thermophilus ribosome (A-and P-site tRNA) bound to BipA-GDPCP | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 3'-amino-3'-deoxyadenosine 5'-(dihydrogen phosphate), ... | | Authors: | Kumar, V, Chen, Y, Ahmed, T, Tan, J, Ero, R, Bhushan, S, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1DT1
 
 | | THERMUS THERMOPHILUS CYTOCHROME C552 SYNTHESIZED BY ESCHERICHIA COLI | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Fee, J.A, Chen, Y, Hill, M.J, Gomez-Moran, E, Loehr, T, Ai, J, Thony-Meyer, L, Williams, P.A, Stura, E, Sridhar, V, McRee, D.E. | | Deposit date: | 2000-01-10 | | Release date: | 2000-02-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Integrity of thermus thermophilus cytochrome c552 synthesized by Escherichia coli cells expressing the host-specific cytochrome c maturation genes, ccmABCDEFGH: biochemical, spectral, and structural characterization of the recombinant protein.
Protein Sci., 9, 2000
|
|
4DKF
 
 | | Crystal Structure of Human Interleukin-34 Bound to FAb2 | | Descriptor: | FAb2 Heavy Chain, FAb2 Light Chain, Interleukin-34, ... | | Authors: | Ma, X, Chen, Y, Stawicki, S, Wu, Y, Bazan, J.F, Starovasnik, M.A. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural Basis for the Dual Recognition of Helical Cytokines IL-34 and CSF-1 by CSF-1R.
Structure, 20, 2012
|
|
1P4E
 
 | | Flpe W330F mutant-DNA Holliday Junction Complex | | Descriptor: | 33-MER, 5'-D(*TP*AP*AP*GP*TP*TP*CP*CP*TP*AP*TP*TP*C)-3', 5'-D(*TP*TP*TP*AP*AP*AP*AP*GP*AP*AP*TP*AP*GP*GP*AP*AP*CP*TP*TP*C)-3', ... | | Authors: | Rice, P.A, Chen, Y. | | Deposit date: | 2003-04-23 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of the conserved Trp330 in Flp-mediated recombination. Functional and structural analysis
J.Biol.Chem., 278, 2003
|
|
4DKE
 
 | | Crystal Structure of Human Interleukin-34 Bound to FAb1.1 | | Descriptor: | FAb1.1 Heavy Chain, FAb1.1 Light Chain, Interleukin-34, ... | | Authors: | Ma, X, Chen, Y, Stawicki, S, Wu, Y, Bazan, J.F, Starovasnik, M.A. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for the Dual Recognition of Helical Cytokines IL-34 and CSF-1 by CSF-1R.
Structure, 20, 2012
|
|
4F11
 
 | | Crystal structure of the extracellular domain of human GABA(B) receptor GBR2 | | Descriptor: | Gamma-aminobutyric acid type B receptor subunit 2 | | Authors: | Geng, Y, Xiong, D, Mosyak, L, Malito, D.L, Kniazeff, J, Chen, Y, Burmakina, S, Quick, M, Bush, M, Javitch, J.A, Pin, J.-P, Fan, Q.R. | | Deposit date: | 2012-05-05 | | Release date: | 2012-06-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure and functional interaction of the extracellular domain of human GABA(B) receptor GBR2.
Nat.Neurosci., 15, 2012
|
|
8FIV
 
 | |
8FIW
 
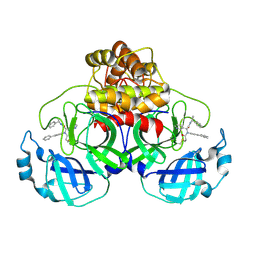 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor Jun10221 | | Descriptor: | 3C-like proteinase nsp5, N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]prop-2-enamide, N-([1,1'-biphenyl]-4-yl)-N-[(1S)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]prop-2-enamide | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2022-12-16 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Exploring diverse reactive warheads for the design of SARS-CoV-2 main protease inhibitors.
Eur.J.Med.Chem., 259, 2023
|
|
1R78
 
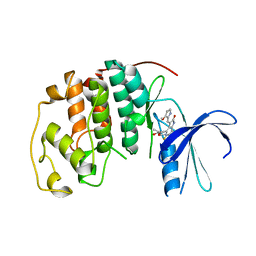 | | CDK2 complex with a 4-alkynyl oxindole inhibitor | | Descriptor: | 4-((3R,4S,5R)-4-AMINO-3,5-DIHYDROXY-HEX-1-YNYL)-5-FLUORO-3-[1-(3-METHOXY-1H-PYRROL-2-YL)-METH-(Z)-YLIDENE]-1,3-DIHYDRO-INDOL-2-ONE, Cell division protein kinase 2 | | Authors: | Luk, K.-C, Simcox, M.E, Schutt, A, Rowan, K, Thompson, T, Chen, Y, Kammlott, U, DePinto, W, Dunten, P, Dermatakis, A. | | Deposit date: | 2003-10-20 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new series of potent oxindole inhibitors of CDK2
Bioorg.Med.Chem.Lett., 14, 2004
|
|
4G72
 
 | |
4G7Q
 
 | |
4F12
 
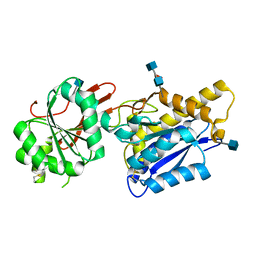 | | Crystal structure of the extracellular domain of human GABA(B) receptor GBR2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 2 | | Authors: | Geng, Y, Xiong, D, Mosyak, L, Malito, D.L, Kniazeff, J, Chen, Y, Burmakina, S, Quick, M, Bush, M, Javitch, J.A, Pin, J.-P, Fan, Q.R. | | Deposit date: | 2012-05-05 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structure and functional interaction of the extracellular domain of human GABA(B) receptor GBR2.
Nat.Neurosci., 15, 2012
|
|
1RYU
 
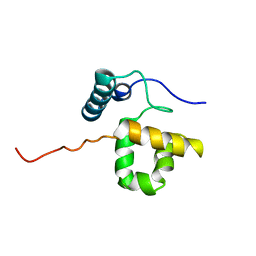 | | Solution Structure of the SWI1 ARID | | Descriptor: | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin subfamily F member 1 | | Authors: | Kim, S, Zhang, Z, Upchurch, S, Isern, N, Chen, Y. | | Deposit date: | 2003-12-22 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and DNA-binding sites of the SWI1 AT-rich interaction domain (ARID) suggest determinants for sequence-specific DNA recognition.
J.Biol.Chem., 279, 2004
|
|
4G7R
 
 | |
4G71
 
 | |
2LAS
 
 | | Molecular Determinants of Paralogue-Specific SUMO-SIM Recognition | | Descriptor: | M-IR2_peptide, Small ubiquitin-related modifier 1 | | Authors: | Namanja, A, Li, Y, Su, Y, Wong, S, Lu, J, Colson, L, Wu, C, Li, S, Chen, Y. | | Deposit date: | 2011-03-20 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into High Affinity Small Ubiquitin-like Modifier (SUMO) Recognition by SUMO-interacting Motifs (SIMs) Revealed by a Combination of NMR and Peptide Array Analysis.
J.Biol.Chem., 287, 2012
|
|
2JWS
 
 | | Solution NMR structures of two designed proteins with 88% sequence identity but different fold and function | | Descriptor: | Ga88 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2007-10-24 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structures of two designed proteins with high sequence identity but different fold and function
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4E0R
 
 | | Structure of the chicken MHC class I molecule BF2*0401 | | Descriptor: | 8-MERIC PEPTIDE (FUS/TLS), Beta-2 microglobulin, MHC class I alpha chain 2 | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-03-05 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
2KDM
 
 | | NMR structures of GA95 and GB95, two designed proteins with 95% sequence identity but different folds and functions | | Descriptor: | DESIGNED PROTEIN | | Authors: | He, Y, Alexander, P, Chen, Y, Bryan, P, Orban, J. | | Deposit date: | 2009-01-12 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From the Cover: A minimal sequence code for switching protein structure and function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3Q2C
 
 | | Binding properties to HLA class I molecules and the structure of the leukocyte Ig-like receptor A3 (LILRA3/ILT6/LIR4/CD85e) | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily A member 3 | | Authors: | Ryu, M, Chen, Y, Qi, J.X, Liu, J, Shi, Y, Cheng, H, Gao, G.F. | | Deposit date: | 2010-12-20 | | Release date: | 2011-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | LILRA3 binds both classical and non-classical HLA class I molecules but with reduced affinities compared to LILRB1/LILRB2: structural evidence
Plos One, 6, 2011
|
|
5XVS
 
 | | Crystal structure of UDP-GlcNAc 2-epimerase NeuC complexed with UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP/UDP-N,N'-diacetylbacillosamine 2-epimerase (Hydrolyzing), LITHIUM ION, ... | | Authors: | Ko, T.P, Hsieh, T.J, Chen, S.C, Wu, S.C, Guan, H.H, Yang, C.H, Chen, C.J, Chen, Y. | | Deposit date: | 2017-06-28 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | The tetrameric structure of sialic acid-synthesizing UDP-GlcNAc 2-epimerase fromAcinetobacter baumannii: A comparative study with human GNE.
J. Biol. Chem., 293, 2018
|
|
2LHC
 
 | | Ga98 solution structure | | Descriptor: | Ga98 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2011-08-08 | | Release date: | 2012-02-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mutational tipping points for switching protein folds and functions.
Structure, 20, 2012
|
|
