8XTR
 
 | | Comamonas testosteroni KF-1 circularly permuted group II intron 2S state | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (219-MER), ... | | Authors: | Wang, L, Xie, J.H, Zhang, C, Zou, J, Huang, Z, Shang, S, Chen, X, Yang, Y, Liu, J, Dong, H, Huang, D, Su, Z. | | Deposit date: | 2024-01-11 | | Release date: | 2025-02-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural basis of circularly permuted group II intron self-splicing.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8YMX
 
 | |
8YMW
 
 | |
3RJW
 
 | | Crystal structure of histone lysine methyltransferase g9a with an inhibitor | | Descriptor: | 2-cyclohexyl-6-methoxy-N-[1-(1-methylethyl)piperidin-4-yl]-7-(3-pyrrolidin-1-ylpropoxy)quinazolin-4-amine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Dong, A, Wasney, G.A, Tempel, W, Liu, F, Barsyte, D, Allali-Hassani, A, Chen, X, Chau, I, Hajian, T, Senisterra, G, Chavda, N, Arora, K, Siarheyeva, A, Kireev, D.B, Herold, J.M, Bochkarev, A, Bountra, C, Weigelt, J, Edwards, A.M, Frye, S.V, Arrowsmith, C.H, Brown, P.J, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-04-15 | | Release date: | 2011-05-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells.
Nat.Chem.Biol., 7, 2011
|
|
3RGA
 
 | | Crystal structure of epoxide hydrolase for polyether lasalocid A biosynthesis | | Descriptor: | (4R,5S)-3-[(2R)-2-{(2S,2'R,4S,5S,5'R)-2,5'-diethyl-5'-[(1S)-1-hydroxyethyl]-4-methyloctahydro-2,2'-bifuran-5-yl}butanoyl]-4-methyl-5-phenyl-1,3-oxazolidin-2-one, (4R,5S)-3-[(2R,3S,4S)-2-ethyl-5-[(3R)-2-ethyl-3-[2-[(2R,3R)-2-ethyl-3-methyl-oxiran-2-yl]ethyl]oxiran-2-yl]-3-hydroxy-4-methyl-pentanoyl]-4-methyl-5-phenyl-1,3-oxazolidin-2-one, ACETATE ION, ... | | Authors: | Hotta, K, Mathews, I.I, Chen, X, Kim, C.-Y. | | Deposit date: | 2011-04-08 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Enzymatic catalysis of anti-Baldwin ring closure in polyether biosynthesis
Nature, 483, 2012
|
|
3S44
 
 | | Crystal Structure of Pasteurella multocida sialyltransferase M144D mutant with CMP bound | | Descriptor: | Alpha-2,3/2,6-sialyltransferase/sialidase, CMP-3F(a)-Neu5Ac | | Authors: | Sugiarto, G, Lau, K, Li, Y, Lim, S, Ames, J.B, Le, D.-T, Fisher, A.J, Chen, X. | | Deposit date: | 2011-05-18 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Sialyltransferase Mutant with Decreased Donor Hydrolysis and Reduced Sialidase Activities for Directly Sialylating Lewis(x).
Acs Chem.Biol., 7, 2012
|
|
9NTJ
 
 | | Chimeric Adenosine deaminase growth factor (ADGF) in complex with pentostatin | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCOFORMYCIN, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kaur, G, Horton, J.R, Cheng, X. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural basis for the substrate specificity of Helix pomatia AMP deaminase and a chimeric ADGF adenosine deaminase.
J.Biol.Chem., 2025
|
|
9NTE
 
 | | Helix pomatia AMP deaminase (HPAMPD) apoenzyme | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kaur, G, Horton, J.R, Cheng, X. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-18 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the substrate specificity of Helix pomatia AMP deaminase and a chimeric ADGF adenosine deaminase.
J.Biol.Chem., 2025
|
|
9NTF
 
 | | Helix pomatia AMP deaminase (HPAMPD) with unknown density in the active site | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kaur, G, Horton, J.R, Cheng, X. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural basis for the substrate specificity of Helix pomatia AMP deaminase and a chimeric ADGF adenosine deaminase.
J.Biol.Chem., 2025
|
|
9NTG
 
 | | Helix pomatia AMP deaminase (HPAMPD) in complex with Pentostatin | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCOFORMYCIN, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Kaur, G, Horton, J.R, Cheng, X. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Helix pomatia AMP deaminase (HPAMPD) in complex with Pentostatin (DCF)
To Be Published
|
|
9NTK
 
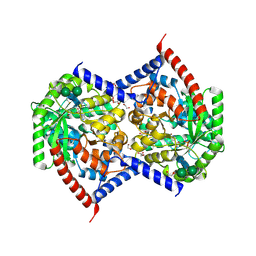 | | Chimeric Adenosine deaminase growth factor (ADGF) in complex with pentostatin monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Adenosine deaminase AGSA,Adenosine deaminase AGSA,Chimeric Adenosine deaminase growth factor, ... | | Authors: | Kaur, G, Horton, J.R, Cheng, X. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-18 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural basis for the substrate specificity of Helix pomatia AMP deaminase and a chimeric ADGF adenosine deaminase.
J.Biol.Chem., 2025
|
|
9NTI
 
 | | Chimeric Adenosine deaminase growth factor (ADGF) apoenzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adenosine deaminase AGSA,Adenosine deaminase AGSA,Chimeric adenosine deaminase growth factor, ZINC ION | | Authors: | Kaur, G, Horton, J.R, Cheng, X. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the substrate specificity of Helix pomatia AMP deaminase and a chimeric ADGF adenosine deaminase.
J.Biol.Chem., 2025
|
|
9NTH
 
 | | Helix pomatia AMP deaminase (HPAMPD) in complex with Pentostatin monophosphate (PMP) | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kaur, G, Horton, J.R, Cheng, X. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-18 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural basis for the substrate specificity of Helix pomatia AMP deaminase and a chimeric ADGF adenosine deaminase.
J.Biol.Chem., 2025
|
|
7VI7
 
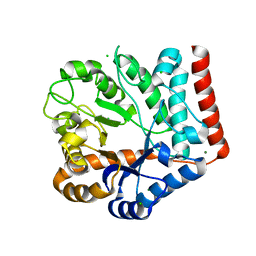 | | Crystal structure of GH3 beta-N-acetylhexosaminidase Amuc_2109 from Akkermansia muciniphila in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Qian, K, Yang, W, Chen, X, Wang, Y, Zhang, M, Wang, M. | | Deposit date: | 2021-09-26 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of a GH3 beta-N-acetylhexosaminidase from Akkermansia muciniphila involved in mucin degradation
Biochem.Biophys.Res.Commun., 589, 2022
|
|
7VI6
 
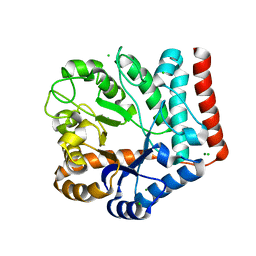 | | Crystal structure of GH3 beta-N-acetylhexosaminidase Amuc_2109 from Akkermansia muciniphila | | Descriptor: | Beta-N-acetylhexosaminidase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Qian, K, Yang, W, Chen, X, Wang, Y, Zhang, M, Wang, M. | | Deposit date: | 2021-09-26 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of a GH3 beta-N-acetylhexosaminidase from Akkermansia muciniphila involved in mucin degradation
Biochem.Biophys.Res.Commun., 589, 2022
|
|
1F0O
 
 | |
1EYU
 
 | |
7CK6
 
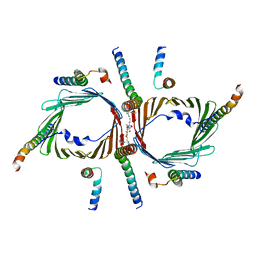 | | Protein translocase of mitochondria | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM40 homolog, ... | | Authors: | Yang, M, Wang, W, Zhang, L, Chen, X. | | Deposit date: | 2020-07-15 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomic structure of human TOM core complex.
Cell Discov, 6, 2020
|
|
7EKA
 
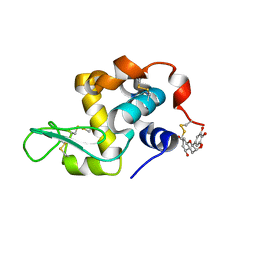 | | crystal structure of epigallocatechin binding with alpha-lactalbumin | | Descriptor: | 2-(3,4,5-TRIHYDROXY-PHENYL)-CHROMAN-3,5,7-TRIOL, Alpha-lactalbumin | | Authors: | Ma, J, Yao, Q, Chen, X, Zang, J. | | Deposit date: | 2021-04-05 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Weak Binding of Epigallocatechin to alpha-Lactalbumin Greatly Improves Its Stability and Uptake by Caco-2 Cells.
J.Agric.Food Chem., 69, 2021
|
|
6OX2
 
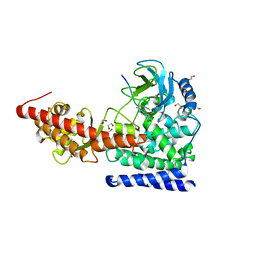 | | SETD3in Complex with an Actin Peptide with the Target Histidine Fully Methylated | | Descriptor: | 1,2-ETHANEDIOL, Actin Peptide, GLYCEROL, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
6OX4
 
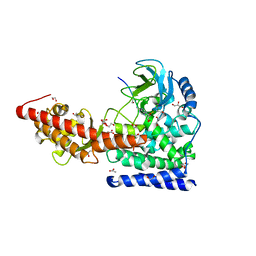 | | A SETD3 Mutant (N255A) in Complex with an Actin Peptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Actin Peptide, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
6OX3
 
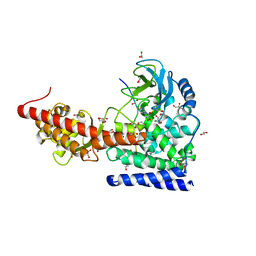 | | SETD3 in Complex with an Actin Peptide with His73 Replaced with Lysine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Actin Peptide, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.785 Å) | | Cite: | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
6OX1
 
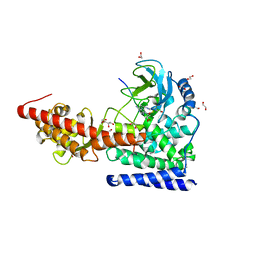 | | SETD3 in Complex with an Actin Peptide with Target Histidine Partially Methylated | | Descriptor: | 1,2-ETHANEDIOL, Actin, cytoplasmic 1, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
6OX5
 
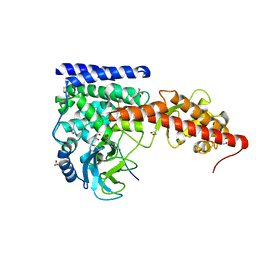 | | A SETD3 Mutant (N255A) in Complex with an Actin Peptide with His73 Replaced with Lysine | | Descriptor: | 1,2-ETHANEDIOL, Actin Peptide, Actin-histidine N-methyltransferase, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
3F8I
 
 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG, crystal structure in space group P21 | | Descriptor: | 5'-D(*DCP*DCP*DAP*DTP*DGP*(5CM)P*DGP*DCP*DTP*DGP*DAP*DC)-3', 5'-D(*DGP*DTP*DCP*DAP*DGP*DCP*DGP*DCP*DAP*DTP*DGP*DG)-3', E3 ubiquitin-protein ligase UHRF1 | | Authors: | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2008-11-12 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
