8VL3
 
 | |
8URE
 
 | |
8VL4
 
 | |
8UTM
 
 | |
3T59
 
 | |
3KO2
 
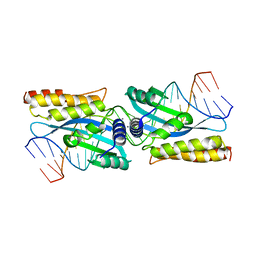 | | I-MsoI re-designed for altered DNA cleavage specificity (-7C) | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*GP*TP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*AP*CP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*CP*CP*G)-3', CALCIUM ION, ... | | Authors: | Taylor, G.K, Stoddard, B.L. | | Deposit date: | 2009-11-13 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs.
Nucleic Acids Res., 38, 2010
|
|
5CY5
 
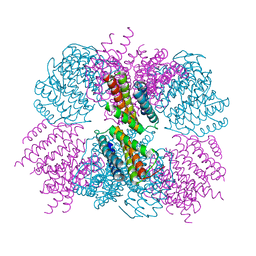 | | Crystal structure of the T33-51H designed self-assembling protein tetrahedron | | Descriptor: | T33-51H-A, T33-51H-B | | Authors: | Cannon, K.A, Cascio, D, Park, R, Boyken, S, King, N, Yeates, T.O. | | Deposit date: | 2015-07-30 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Design and structure of two new protein cages illustrate successes and ongoing challenges in protein engineering.
Protein Sci., 29, 2020
|
|
7UNJ
 
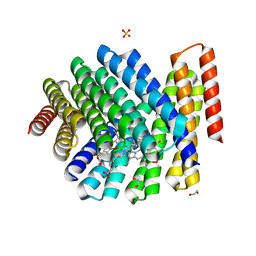 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester matching geometry of purple bacterial special pair, SP1-ZnPPaM | | Descriptor: | 1,2-ETHANEDIOL, SP1-ZnPPaM designed chlorophyll dimer protein, SULFATE ION, ... | | Authors: | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 2024
|
|
7UNH
 
 | |
7UNI
 
 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester, SP2-ZnPPaM | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, SP2-ZnPPaM designed chlorophyll dimer protein, ... | | Authors: | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 2024
|
|
7UZL
 
 | |
6P6F
 
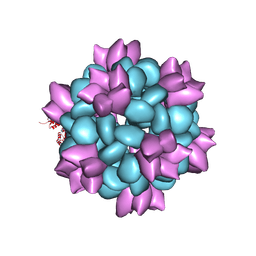 | | BG505 SOSIP-I53-50NP | | Descriptor: | I53-50A.1NT1, I53-50B.4PT1 | | Authors: | Berndsen, Z.T, Ward, A.B. | | Deposit date: | 2019-06-03 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Enhancing and shaping the immunogenicity of native-like HIV-1 envelope trimers with a two-component protein nanoparticle.
Nat Commun, 10, 2019
|
|
8CTO
 
 | |
8CUN
 
 | |
8CWA
 
 | |
7UBH
 
 | |
4F2V
 
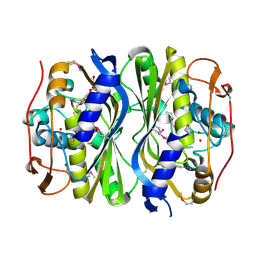 | | Crystal Structure of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR165 | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-ALPHA-D-MALTOSIDE, De novo designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Rajagopalan, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
3Q6O
 
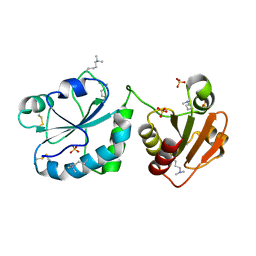 | | Oxidoreductase Fragment of Human QSOX1 | | Descriptor: | SULFATE ION, Sulfhydryl oxidase 1 | | Authors: | Fass, D, Alon, A. | | Deposit date: | 2011-01-03 | | Release date: | 2012-05-30 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The dynamic disulphide relay of quiescin sulphydryl oxidase.
Nature, 488, 2012
|
|
3QD9
 
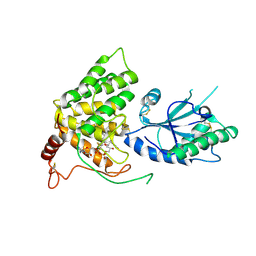 | |
3LEF
 
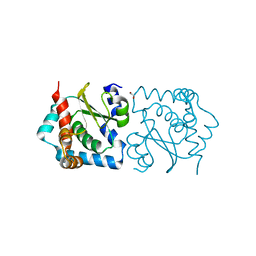 | | Crystal structure of HIV epitope-scaffold 4E10_S0_1Z6NA_001 | | Descriptor: | 1,2-ETHANEDIOL, Uncharacterized protein 4E10_S0_1Z6NA_001 (T18) | | Authors: | Holmes, M.A. | | Deposit date: | 2010-01-14 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational Design of Epitope-Scaffolds Allows Induction of Antibodies Specific for a Poorly Immunogenic HIV Vaccine Epitope.
Structure, 18, 2010
|
|
7Q1V
 
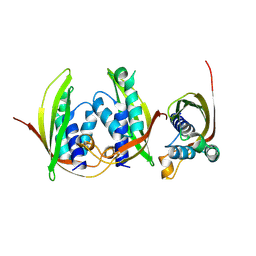 | |
3LHP
 
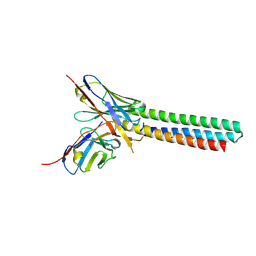 | |
3LH2
 
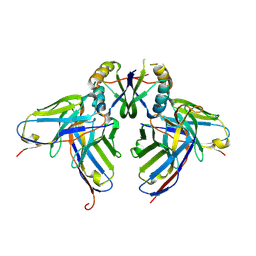 | |
3LF9
 
 | | Crystal structure of HIV epitope-scaffold 4E10_D0_1IS1A_001_C | | Descriptor: | 4E10_D0_1IS1A_001_C (T161) | | Authors: | Holmes, M.A. | | Deposit date: | 2010-01-16 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational Design of Epitope-Scaffolds Allows Induction of Antibodies Specific for a Poorly Immunogenic HIV Vaccine Epitope.
Structure, 18, 2010
|
|
3PPD
 
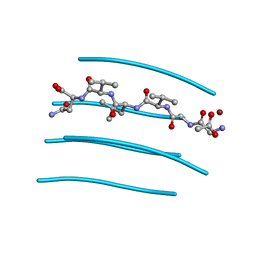 | | GGVLVN segment from Human Prostatic Acid Phosphatase Residues 260-265, involved in Semen-Derived Enhancer of Viral Infection | | Descriptor: | ACETIC ACID, GGVLVN peptide, amyloid forming segment, ... | | Authors: | Zhao, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-11-24 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation.
Nature, 475, 2011
|
|
