6PH4
 
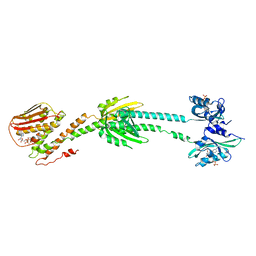 | | Full length LOV-PAS-HK construct from the LOV-HK sensory protein from Brucella abortus (light-adapted, construct 15-489) | | Descriptor: | Blue-light-activated histidine kinase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Rinaldi, J, Otero, L.H, Fernandez, I, Goldbaum, F.A, Shin, H, Yang, X, Klinke, S. | | Deposit date: | 2019-06-25 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Dimer Asymmetry and Light Activation Mechanism in Brucella Blue-Light Sensor Histidine Kinase.
Mbio, 12, 2021
|
|
3EKD
 
 | |
3W9D
 
 | |
3F2P
 
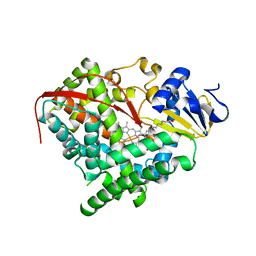
 | | Thermolysin inhibition | | Descriptor: | 3-methyl-2-(propanoyloxy)benzoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Englert, L, Heine, A, Klebe, G. | | Deposit date: | 2008-10-30 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-Based Lead Discovery: Screening and Optimizing Fragments for Thermolysin Inhibition.
Chemmedchem, 5, 2010
|
|
3FCQ
 
 | | Thermolysin inhibition | | Descriptor: | 2-(acetyloxy)-3-methylbenzoic acid, CALCIUM ION, Thermolysin, ... | | Authors: | Steuber, H, Englert, L, Silber, K, Heine, A, Klebe, G. | | Deposit date: | 2008-11-22 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fragment-Based Lead Discovery: Screening and Optimizing Fragments for Thermolysin Inhibition.
Chemmedchem, 5, 2010
|
|
3F28
 
 | | Thermolysin inhibition | | Descriptor: | 2-[(cyclopropylcarbonyl)oxy]-3-methylbenzoic acid, CALCIUM ION, Thermolysin, ... | | Authors: | Englert, L, Heine, A, Klebe, G. | | Deposit date: | 2008-10-29 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Fragment-Based Lead Discovery: Screening and Optimizing Fragments for Thermolysin Inhibition.
Chemmedchem, 5, 2010
|
|
3CXV
 
 | |
3W9E
 
 | | Structure of Human Monoclonal Antibody E317 Fab Complex with HSV-2 gD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab heavy chain, Antibody Fab light chain, ... | | Authors: | Lee, C.C, Lin, L.L, Wang, A.H.J. | | Deposit date: | 2013-04-03 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the antibody neutralization of herpes simplex virus
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3E4W
 
 | |
3EKB
 
 | |
3ERR
 
 | |
3GD8
 
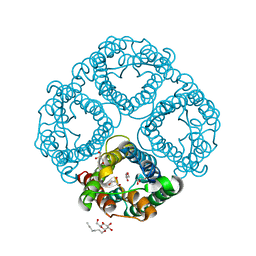 | | Crystal Structure of Human Aquaporin 4 at 1.8 and its Mechanism of Conductance | | Descriptor: | Aquaporin-4, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Ho, J.D, Yeh, R, Sandstrom, A, Chorny, I, Harries, W.E.C, Robbins, R.A, Miercke, L.J.W, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2009-02-23 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human aquaporin 4 at 1.8 A and its mechanism of conductance.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1LX6
 
 | |
8TKM
 
 | | Murine NF-kappaB p50 Rel Homology Region homodimer in complex with 17-mer kappaB DNA from human interleukin-6 (IL-6) promoter | | Descriptor: | 17-mer kappaB DNA, Nuclear factor NF-kappa-B p50 subunit | | Authors: | Zhu, N, Mealka, M, Mitchel, S, Rogers, W.E, Huxford, T. | | Deposit date: | 2023-07-25 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Crystallographic Study of Preferred Spacing by the NF-kappa B p50 Homodimer on kappa B DNA.
Biomolecules, 13, 2023
|
|
8TKN
 
 | | Murine NF-kappaB p50 Rel Homology Region homodimer in complex with 10-mer kappaB DNA from human Neutrophil Gelatinase-associated Lipocalin (NGAL) promoter | | Descriptor: | DNA A, DNA B, Nuclear factor NF-kappa-B p50 subunit | | Authors: | Zhu, N, Mealka, M, Mitchel, S, Rogers, W.E, Huxford, T. | | Deposit date: | 2023-07-25 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Crystallographic Study of Preferred Spacing by the NF-kappa B p50 Homodimer on kappa B DNA.
Biomolecules, 13, 2023
|
|
8TKL
 
 | | Murine NF-kappaB p50 Rel Homology Region homodimer in complex with a Test 16-mer kappaB-like DNA | | Descriptor: | Nuclear factor NF-kappa-B p50 subunit, Test 17-mer kappaB-like DNA | | Authors: | Mitchel, S, Mealka, M, Rogers, W.E, Milani, C, Acuna, L.M, Huxford, T. | | Deposit date: | 2023-07-25 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray Crystallographic Study of Preferred Spacing by the NF-kappa B p50 Homodimer on kappa B DNA.
Biomolecules, 13, 2023
|
|
5LBV
 
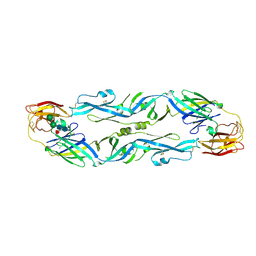 | | Structural basis of zika and dengue virus potent antibody cross-neutralization | | Descriptor: | SODIUM ION, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, envelope protein E | | Authors: | Barba-Spaeth, G. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of potent Zika-dengue virus antibody cross-neutralization.
Nature, 536, 2016
|
|
5LBS
 
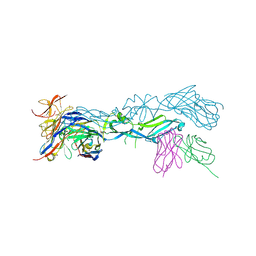 | | structural basis of Zika and Dengue virus potent antibody cross-neutralization | | Descriptor: | 1,2-ETHANEDIOL, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C8, SULFATE ION, ... | | Authors: | Vaney, M.C, Rouvinski, A, Barba-Spaeth, G, Rey, F.A. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis of potent Zika-dengue virus antibody cross-neutralization.
Nature, 536, 2016
|
|
1VJE
 
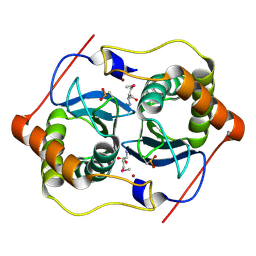 | |
1O63
 
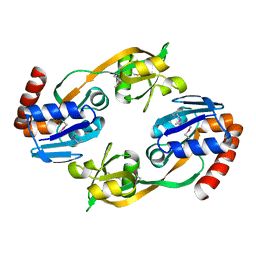 | |
1O6C
 
 | |
4G57
 
 | | Staphylococcal Nuclease double mutant I72L, I92L | | Descriptor: | Thermonuclease | | Authors: | Sakon, J, Stites, W.E, Gill, E, Latimer, E, Rosser, J. | | Deposit date: | 2012-07-17 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: |
|
|
1O66
 
 | |
1O60
 
 | | Crystal structure of KDO-8-phosphate synthase | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O67
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | Hypothetical protein yiiM | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
