6TYF
 
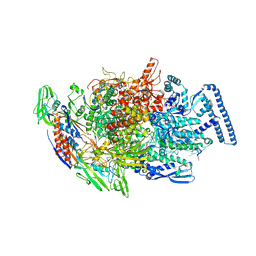 | | Crystal structure of MTB sigma L transcription initiation complex with 6 nt long RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*AP*TP*CP*GP*AP*GP*GP*GP*T)-3'), DNA (5'-D(P*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2019-08-08 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7RVA
 
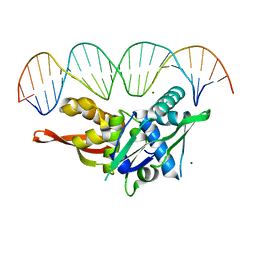 | | Updated Crystal Structure of Replication Initiator Protein REPE54. | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*T)-3'), DNA (5'-D(*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*T)-3'), MAGNESIUM ION, ... | | Authors: | Ward, A.R, Snow, C.D. | | Deposit date: | 2021-08-18 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Stabilizing DNA-Protein Co-Crystals via Intra-Crystal Chemical Ligation of the DNA
Crystals, 12, 2022
|
|
6TYG
 
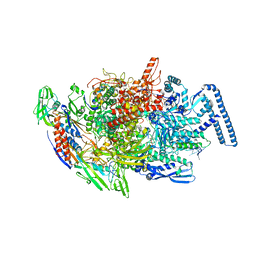 | | Crystal structure of MTB sigma L transcription initiation complex with 9 nt long RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*AP*TP*CP*GP*AP*GP*GP*GP*TP*G)-3'), DNA (5'-D(P*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2019-08-08 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8TIX
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 1 sticky bases and 5' terminal phosphates and crosslinked with EDC. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3'), DNA (5'-D(A*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TIR
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 1 sticky bases and 5' terminal phosphates. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3'), DNA (5'-D(A*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TIU
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 2 sticky base overhangs and 3' terminal phosphates. | | Descriptor: | DNA (5'-D(CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*GP*A)-3')|, DNA (5'-D(GP*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*A)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TIS
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 1 sticky bases and 3' terminal phosphates. | | Descriptor: | DNA (5'-D(GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*GP*A)-3'), DNA(5'-D(CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*GP*A)-')|, MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TIW
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with blunt ends and 3' terminal phosphates and crosslinked with EDC. | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*GP*A)-3', DNA (5'-D(*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*GP*GP*A)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TIV
 
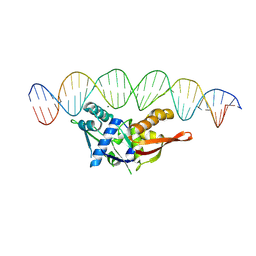 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with blunt ends and 5' terminal phosphates and crosslinked with EDC. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3')|, DNA (5'-D(A*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TJ1
 
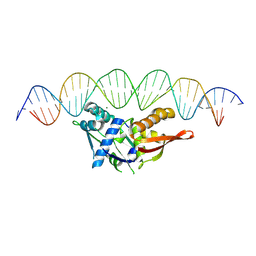 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 2 sticky base overhangs and 3' terminal phosphates and crosslinked with EDC. | | Descriptor: | DNA (5'-D(CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*GP*A)-3')|, DNA (5'-D(GP*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*A)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TIQ
 
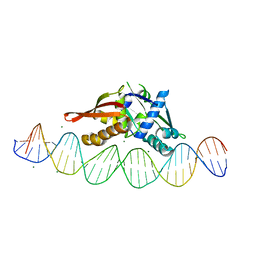 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with blunt ends and 3' terminal phosphates. | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*GP*A)-3', DNA (5'-D(*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*GP*GP*A)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Slaughter, C.K, Shrestha, R, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TIT
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 2 sticky base overhangs and no terminal phosphates. | | Descriptor: | DNA (5'-D(CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3')|, DNA (5'-D(GP*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TIZ
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 2 sticky base overhangs and no terminal phosphates and crosslinked with EDC. | | Descriptor: | DNA (5'-D(CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3')|, DNA (5'-D(GP*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TJ0
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 2 sticky base overhangs and 5' terminal phosphates and crosslinked with EDC. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3'), DNA (5'-D(A*GP*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Slaughter, C.K, Shrestha, R, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TIP
 
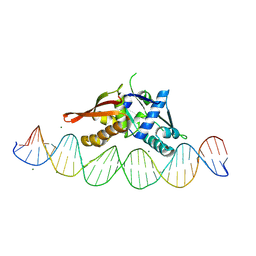 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with blunt ends and 5' terminal phosphates. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3')|, DNA (5'-D(A*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Slaughter, C.K, Shrestha, R, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
8TIY
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 1 sticky bases and 3' terminal phosphates and crosslinked with EDC. | | Descriptor: | DNA (5'-D(GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*GP*A)-3'), DNA(5'-D(CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*GP*A)-')|, MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Tuning Chemical DNA Ligation within DNA Crystals and Protein-DNA Cocrystals.
Acs Nanosci Au, 4, 2024
|
|
7SDP
 
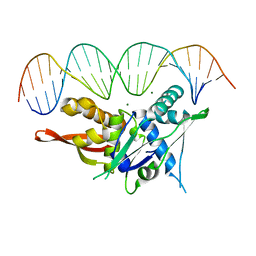 | |
6U1V
 
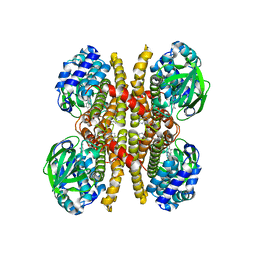 | | Crystal structure of acyl-ACP/acyl-CoA dehydrogenase from allylmalonyl-CoA and FK506 biosynthesis, TcsD | | Descriptor: | Acyl-CoA dehydrogenase domain-containing protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | Authors: | Blake-Hedges, J.M, Pereira, J.H, Barajas, J.F, Adams, P.D, Keasling, J.D. | | Deposit date: | 2019-08-16 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Mechanism of Regioselectivity in an Unusual Bacterial Acyl-CoA Dehydrogenase.
J.Am.Chem.Soc., 142, 2020
|
|
6ULX
 
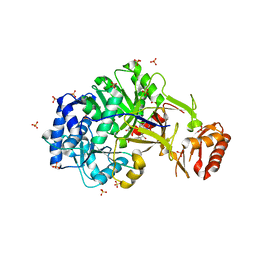 | |
8TBW
 
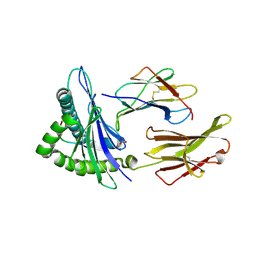 | | Human Class I MHC HLA-A2 in complex with sorting nexin 24 (127-135) peptide KLSHQPVLL | | Descriptor: | Beta-2-microglobulin, HLA-A*02:01 alpha chain, NITRATE ION, ... | | Authors: | Arbuiso, A, Weiss, L.I, Brambley, C.A, Ma, J, Keller, G.L.J, Ayres, C.M, Baker, B.M. | | Deposit date: | 2023-06-29 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Accurate modeling of peptide-MHC structures with AlphaFold.
Structure, 32, 2024
|
|
8TBV
 
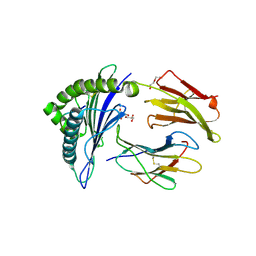 | | Human Class I MHC HLA-A2 in complex with sorting nexin 24 (127-135) neoantigen KLSHQLVLL | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Arbuiso, A, Weiss, L.I, Brambley, C.A, Ma, J, Keller, G.L.J, Ayres, C.M, Baker, B.M. | | Deposit date: | 2023-06-29 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Accurate modeling of peptide-MHC structures with AlphaFold.
Structure, 32, 2024
|
|
6TYE
 
 | | Crystal structure of MTB sigma L transcription initiation complex with 5 nt long RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*AP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(P*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2019-08-08 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8TJG
 
 | |
6MF2
 
 | | Improved Model of Human Coagulation Factor VIII | | Descriptor: | CALCIUM ION, COPPER (I) ION, Coagulation factor VIII, ... | | Authors: | Smith, I.W, Spiegel, P.C. | | Deposit date: | 2018-09-08 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.609364 Å) | | Cite: | The 3.2 angstrom structure of a bioengineered variant of blood coagulation factor VIII indicates two conformations of the C2 domain.
J.Thromb.Haemost., 18, 2020
|
|
8C3P
 
 | |
