5X7S
 
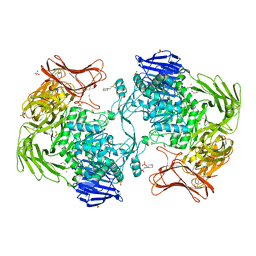 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase, terbium derivative | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
3M63
 
 | |
2Z5V
 
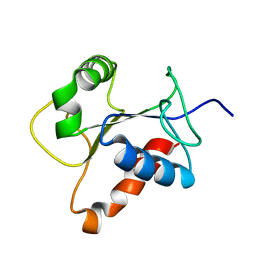 | | Solution structure of the TIR domain of human MyD88 | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Ohnishi, H, Tochio, H, Hiroaki, H, Kondo, N, Kato, Z, Shirakawa, M. | | Deposit date: | 2007-07-19 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the multiple interactions of the MyD88 TIR domain in TLR4 signaling.
Proc.Natl.Acad.Sci.USA, 2009
|
|
5X7Q
 
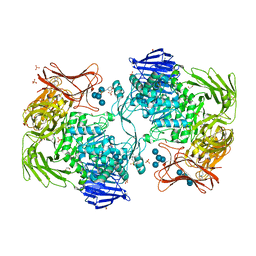 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase complexed with maltohexaose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
3O0T
 
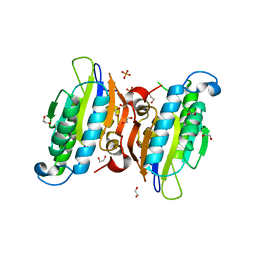 | | Crystal structure of human phosphoglycerate mutase family member 5 (PGAM5) in complex with phosphate | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Serine/threonine-protein phosphatase PGAM5, ... | | Authors: | Chaikuad, A, Alfano, I, Picaud, S, Filippakopoulos, P, Barr, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Takeda, K, Ichijo, H, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-20 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of PGAM5 Provide Insight into Active Site Plasticity and Multimeric Assembly.
Structure, 25, 2017
|
|
4NNJ
 
 | |
1WMN
 
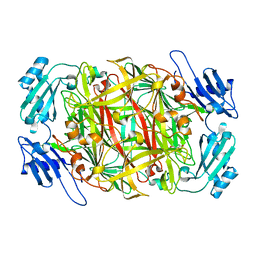 | | Crystal structure of topaquinone-containing amine oxidase activated by cobalt ion | | Descriptor: | COBALT (II) ION, Phenylethylamine oxidase | | Authors: | Okajima, T, Kishishita, S, Chiu, Y.C, Murakawa, T, Kim, M, Yamaguchi, H, Hirota, S, Kuroda, S, Tanizawa, K. | | Deposit date: | 2004-07-13 | | Release date: | 2005-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reinvestigation of metal ion specificity for quinone cofactor biogenesis in bacterial copper amine oxidase
Biochemistry, 44, 2005
|
|
1WSV
 
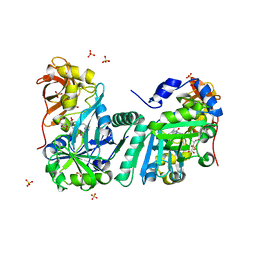 | | Crystal Structure of Human T-protein of Glycine Cleavage System | | Descriptor: | Aminomethyltransferase, N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, SULFATE ION | | Authors: | Okamura-Ikeda, K, Hosaka, H, Yoshimura, M, Yamashita, E, Toma, S, Nakagawa, A, Fujiwara, K, Motokawa, Y, Taniguchi, H. | | Deposit date: | 2004-11-11 | | Release date: | 2005-08-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Human T-protein of Glycine Cleavage System at 2.0A Resolution and its Implication for Understanding Non-ketotic Hyperglycinemia
J.Mol.Biol., 351, 2005
|
|
1SOX
 
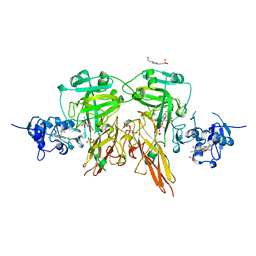 | | SULFITE OXIDASE FROM CHICKEN LIVER | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, MOLYBDENUM ATOM, ... | | Authors: | Kisker, C, Schindelin, H, Rees, D.C. | | Deposit date: | 1997-12-31 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of sulfite oxidase deficiency from the structure of sulfite oxidase.
Cell(Cambridge,Mass.), 91, 1997
|
|
5H41
 
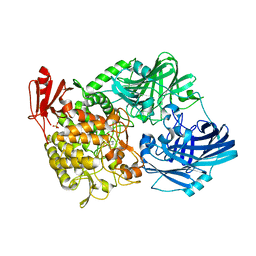 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with sophorose, isofagomine, sulfate ion | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, SULFATE ION, Uncharacterized protein, ... | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
5H40
 
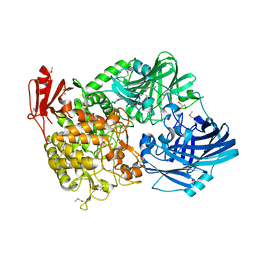 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with sophorose | | Descriptor: | CALCIUM ION, GLYCEROL, Uncharacterized protein, ... | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
1WSR
 
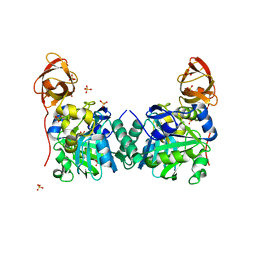 | | Crystal Structure of Human T-protein of Glycine Cleavage System | | Descriptor: | Aminomethyltransferase, SULFATE ION | | Authors: | Okamura-Ikeda, K, Hosaka, H, Yoshimura, M, Yamashita, E, Toma, S, Nakagawa, A, Fujiwara, K, Motokawa, Y, Taniguchi, H. | | Deposit date: | 2004-11-10 | | Release date: | 2005-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human T-protein of Glycine Cleavage System at 2.0A Resolution and its Implication for Understanding Non-ketotic Hyperglycinemia
J.Mol.Biol., 351, 2005
|
|
5H3Z
 
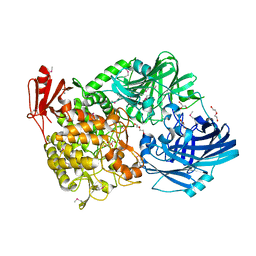 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
5YSD
 
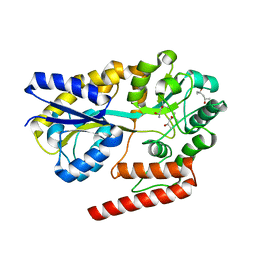 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotriose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
1THJ
 
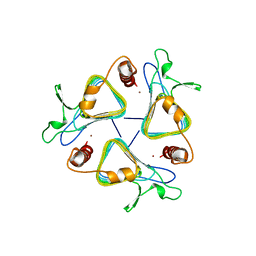 | | CARBONIC ANHYDRASE FROM METHANOSARCINA | | Descriptor: | CARBONIC ANHYDRASE, ZINC ION | | Authors: | Kisker, C, Schindelin, H, Rees, D.C. | | Deposit date: | 1996-04-02 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A left-hand beta-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila.
EMBO J., 15, 1996
|
|
5H42
 
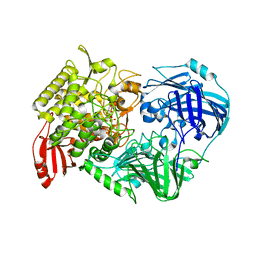 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with alpha-d-glucose-1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, Uncharacterized protein, alpha-D-glucopyranose | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
5YSF
 
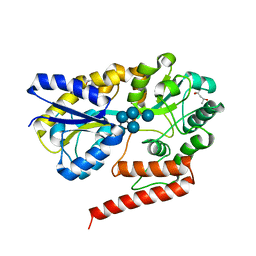 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophoropentaose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSE
 
 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotetraose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSB
 
 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in ligand-free form | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lin1841 protein, ZINC ION | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-13 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
1VFI
 
 | | Solution Structure of Vanabin2 (RUH-017), a Vanadium-binding Protein from Ascidia sydneiensis samea | | Descriptor: | vanadium-binding protein 2 | | Authors: | Hamada, T, Hirota, H, Asanuma, M, Hayashi, F, Kobayashi, N, Ueki, T, Michibata, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-04-13 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Vanabin2, a Vanadium(IV)-Binding Protein from the Vanadium-Rich Ascidian Ascidia sydneiensis samea
J.Am.Chem.Soc., 127, 2005
|
|
1UFB
 
 | | Crystal structure of TT1696 from Thermus thermophilus HB8 | | Descriptor: | TT1696 protein | | Authors: | Idaka, M, Murayama, K, Yamaguchi, H, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-28 | | Release date: | 2003-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of TT1696 from Thermus thermophilus HB8
To be published
|
|
3VUZ
 
 | | Crystal structure of histone methyltransferase SET7/9 in complex with AAM-1 | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](hexyl)amino}-5'-deoxyadenosine, Histone-lysine N-methyltransferase SETD7 | | Authors: | Niwa, H, Handa, N, Tomabechi, Y, Honda, K, Toyama, M, Ohsawa, N, Shirouzu, M, Kagechika, H, Hirano, T, Umehara, T, Yokoyama, S. | | Deposit date: | 2012-07-10 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of histone methyltransferase SET7/9 in complexes with adenosylmethionine derivatives
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1TV8
 
 | | Structure of MoaA in complex with S-adenosylmethionine | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, IRON/SULFUR CLUSTER, Molybdenum cofactor biosynthesis protein A, ... | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2004-06-28 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
3QQ7
 
 | |
1UFA
 
 | | Crystal structure of TT1467 from Thermus thermophilus HB8 | | Descriptor: | TT1467 protein | | Authors: | Idaka, M, Terada, T, Murayama, K, Yamaguchi, H, Nureki, O, Ishitani, R, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-28 | | Release date: | 2003-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of TT1467 from Thermus thermophilus HB8
To be published
|
|
