4GPL
 
 | |
7XVG
 
 | | Cryo-EM structure of binary complex of plant NLR Sr35 and effector AvrSr35 | | Descriptor: | AvrSr35, Sr35 | | Authors: | Ouyang, S.Y, Zhao, Y.B, Li, Z.K, Liu, M.X. | | Deposit date: | 2022-05-23 | | Release date: | 2022-09-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Pathogen effector AvrSr35 triggers Sr35 resistosome assembly via a direct recognition mechanism.
Sci Adv, 8, 2022
|
|
7Y3E
 
 | |
7DE1
 
 | |
5J8V
 
 | |
7V6L
 
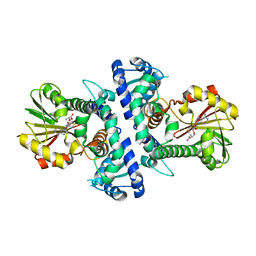 | | LcCOMT in complex with SAH | | Descriptor: | LcCOMT, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yu, Y, CHen, Q. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Structure basis of the caffeic acid O-methyltransferase from Ligusiticum chuanxiong to understand its selective mechanism.
Int.J.Biol.Macromol., 194, 2022
|
|
7V6J
 
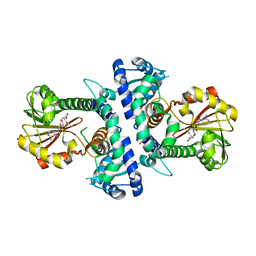 | | LcCOMT in complex with SAM | | Descriptor: | LcCOMT, S-ADENOSYLMETHIONINE, SODIUM ION | | Authors: | Yu, Y, CHen, Q. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structure basis of the caffeic acid O-methyltransferase from Ligusiticum chuanxiong to understand its selective mechanism.
Int.J.Biol.Macromol., 194, 2022
|
|
6J8R
 
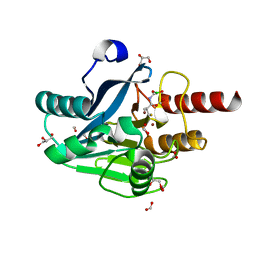 | | Metallo-Beta-Lactamase VIM-2 in complex with Dual MBL/SBL Inhibitor MS01 | | Descriptor: | Beta-lactamase class B VIM-2, FORMIC ACID, GLYCEROL, ... | | Authors: | Li, G.-B, Liu, S. | | Deposit date: | 2019-01-21 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.575 Å) | | Cite: | Structure-Based Development of (1-(3'-Mercaptopropanamido)methyl)boronic Acid Derived Broad-Spectrum, Dual-Action Inhibitors of Metallo- and Serine-beta-lactamases.
J.Med.Chem., 62, 2019
|
|
6J8Q
 
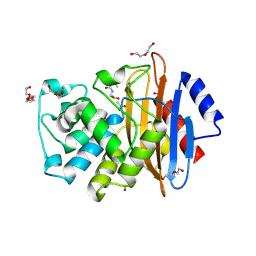 | | Serine Beta-Lactamase KPC-2 in Complex with Dual MBL/SBL Inhibitor WL-001 | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Li, G.-B, Liu, S. | | Deposit date: | 2019-01-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | Structure-Based Development of (1-(3'-Mercaptopropanamido)methyl)boronic Acid Derived Broad-Spectrum, Dual-Action Inhibitors of Metallo- and Serine-beta-lactamases.
J.Med.Chem., 62, 2019
|
|
6JN4
 
 | | Serine Beta-Lactamase KPC-2 in Complex with Dual MBL/SBL Inhibitor WL-001 | | Descriptor: | ACETATE ION, ACETIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, G.-B, Liu, S. | | Deposit date: | 2019-03-13 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Development of (1-(3'-Mercaptopropanamido)methyl)boronic Acid Derived Broad-Spectrum, Dual-Action Inhibitors of Metallo- and Serine-beta-lactamases.
J.Med.Chem., 62, 2019
|
|
6JN5
 
 | | Serine Beta-Lactamase KPC-2 in Complex with Dual MBL/SBL Inhibitor MS23 | | Descriptor: | Serine Beta-Lactamase KPC-2, [(S)-(4-fluorophenyl)-[[(2S)-2-methyl-3-sulfanyl-propanoyl]amino]methyl]boronic acid | | Authors: | Li, G.-B, Liu, S. | | Deposit date: | 2019-03-13 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure-Based Development of (1-(3'-Mercaptopropanamido)methyl)boronic Acid Derived Broad-Spectrum, Dual-Action Inhibitors of Metallo- and Serine-beta-lactamases.
J.Med.Chem., 62, 2019
|
|
6JN3
 
 | | Serine Beta-Lactamase KPC-2 in Complex with Dual MBL/SBL Inhibitor MS05 | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, Serine Beta-Lactamase KPC-2, ... | | Authors: | Li, G.-B, Liu, S. | | Deposit date: | 2019-03-13 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.216 Å) | | Cite: | Structure-Based Development of (1-(3'-Mercaptopropanamido)methyl)boronic Acid Derived Broad-Spectrum, Dual-Action Inhibitors of Metallo- and Serine-beta-lactamases.
J.Med.Chem., 62, 2019
|
|
6JN6
 
 | | Metallo-Beta-Lactamase VIM-2 in complex with Dual MBL/SBL Inhibitor MS19 | | Descriptor: | Beta-lactamase class B VIM-2, FORMIC ACID, ZINC ION, ... | | Authors: | Li, G.-B, Liu, S. | | Deposit date: | 2019-03-13 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Structure-Based Development of (1-(3'-Mercaptopropanamido)methyl)boronic Acid Derived Broad-Spectrum, Dual-Action Inhibitors of Metallo- and Serine-beta-lactamases.
J.Med.Chem., 62, 2019
|
|
8WWF
 
 | |
8WWE
 
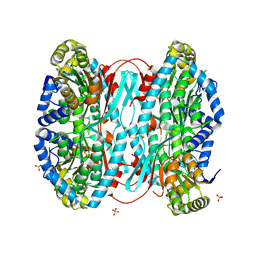 | |
8UG4
 
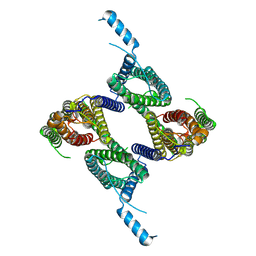 | |
8WWD
 
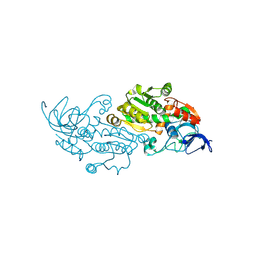 | |
8UG7
 
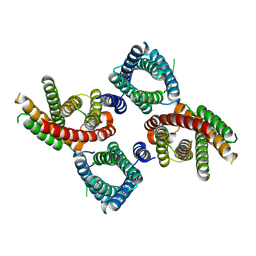 | | Mus musculus Otopetrin 2 (mOTOP2) in pH 8.0 | | Descriptor: | Proton channel OTOP2 | | Authors: | Gan, N, Jiang, Y. | | Deposit date: | 2023-10-05 | | Release date: | 2024-08-21 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural mechanism of proton conduction in otopetrin proton channel.
Nat Commun, 15, 2024
|
|
8UG5
 
 | |
8UGA
 
 | |
8UG6
 
 | | Mus musculus Otopetrin 2 (mOTOP2) in pH 5.0 | | Descriptor: | Proton channel OTOP2 | | Authors: | Gan, N, Jiang, Y. | | Deposit date: | 2023-10-05 | | Release date: | 2024-08-21 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural mechanism of proton conduction in otopetrin proton channel.
Nat Commun, 15, 2024
|
|
8UG8
 
 | |
8WX8
 
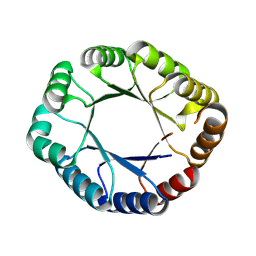 | | De novo design protein -T09 | | Descriptor: | De novo design protein -T09 | | Authors: | Wang, C, Wang, S, Liu, Y. | | Deposit date: | 2023-10-27 | | Release date: | 2024-10-09 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8WWC
 
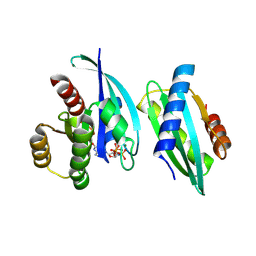 | | De novo design binder of HRAS -120-4 | | Descriptor: | De novo design protein 120-4, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Wang, L, Wang, S, Liu, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-10-09 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8XTV
 
 | | Crystal structure of AaHPPD-Y14150 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, [3-ethyl-1-methyl-2,2-bis(oxidanylidene)-2$l^{6},1,3-benzothiadiazol-5-yl]-(1-methyl-5-oxidanyl-pyrazol-4-yl)methanone | | Authors: | Dong, J, Lin, H.-Y, Yang, G.-F. | | Deposit date: | 2024-01-11 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Repurposing 4-Hydroxyphenylpyruvate dioxygenase inhibitors as novel agents for mosquito control: A structure-based design approach.
Int.J.Biol.Macromol., 315, 2025
|
|
