6W9A
 
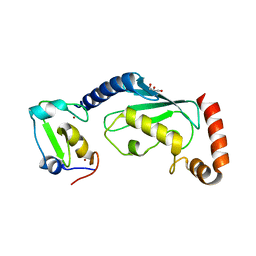 | | RNF12 RING domain in complex with Ube2e2 | | Descriptor: | E3 ubiquitin-protein ligase RLIM, GLYCEROL, Ubiquitin-conjugating enzyme E2 E2, ... | | Authors: | Middleton, A.J, Day, C.L. | | Deposit date: | 2020-03-22 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The RING Domain of RING Finger 12 Efficiently Builds Degradative Ubiquitin Chains.
J.Mol.Biol., 432, 2020
|
|
7F02
 
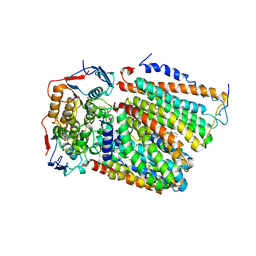 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7F04
 
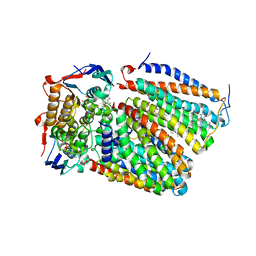 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with Heme and ATP. | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7F03
 
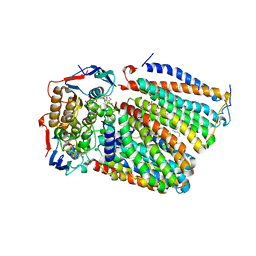 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with ANP | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
8HDS
 
 | | Cyanophage Pam3 portal-adaptor | | Descriptor: | Pam3 adaptor protein, Pam3 portal protein | | Authors: | Yang, F, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-11-06 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Fine structure and assembly pattern of a minimal myophage Pam3.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HDT
 
 | |
8HDW
 
 | | Cyanophage Pam3 Sheath-tube | | Descriptor: | Pam3 sheath protein, pam3 tube | | Authors: | Yang, F, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-11-06 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Fine structure and assembly pattern of a minimal myophage Pam3.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HDR
 
 | | Cyanophage Pam3 neck | | Descriptor: | Pam3 adaptor protein, Pam3 connector protein, Pam3 sheath protein, ... | | Authors: | Yang, F, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-11-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Fine structure and assembly pattern of a minimal myophage Pam3.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6A52
 
 | | Oxidase ChaP-H1 | | Descriptor: | FE (II) ION, dioxidase ChaP-H1 | | Authors: | Zhang, B, Ge, H.M. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
6A5F
 
 | |
8WYP
 
 | |
6A4X
 
 | | Oxidase ChaP-H2 | | Descriptor: | Bleomycin resistance protein, FE (II) ION | | Authors: | Zhang, B, Wang, Y.S, Ge, H.M. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
6A4Z
 
 | | Oxidase ChaP | | Descriptor: | ChaP protein, FE (II) ION | | Authors: | Zhang, B, Ge, H.M. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Basis for the Final Oxidative Rearrangement Steps in Chartreusin Biosynthesis.
J. Am. Chem. Soc., 140, 2018
|
|
6A5G
 
 | |
6A5H
 
 | |
8IYX
 
 | | Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365 | | Descriptor: | 1-[4-(3-chlorophenyl)phenyl]carbonyl-4-[2-(4-phenylmethoxyphenyl)ethanoylamino]piperidine-4-carboxylic acid, Probable G-protein coupled receptor 34,Probable G-protein coupled receptor 34,YL-365 | | Authors: | Jia, G.W, Wang, X, Zhang, C.B, Dong, H.H, Su, Z.M. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6LY4
 
 | | The crystal structure of the BM3 mutant LG-23 in complex with testosterone | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, IMIDAZOLE, ... | | Authors: | Peng, Y, Chen, J, Zhou, J, Li, A, ReetZ, M.T. | | Deposit date: | 2020-02-13 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Regio- and Stereoselective Steroid Hydroxylation at C7 by Cytochrome P450 Monooxygenase Mutants.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5QBV
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, N-[2-chloro-5-(1-{3-[4-(6-chloro-3-methyl-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl)piperidin-1-yl]propyl}-6-oxo-1,6-dihydropyridazin-3-yl)benzyl]benzamide | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Diazinones as P2 replacements for pyrazole-based cathepsin S inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7FE0
 
 | |
5QBY
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, N-[2-chloro-5-(1-{3-[4-(6-chloro-3-methyl-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl)piperidin-1-yl]propyl}-6-oxo-1,6-dihydropyrimidin-5-yl)benzyl]-4-fluorobenzamide | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Diazinones as P2 replacements for pyrazole-based cathepsin S inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6CE0
 
 | | Crystal structure of a HIV-1 clade B tier-3 isolate H078.14 UFO-BG Env trimer in complex with broadly neutralizing Fabs PGT124 and 35O22 at 4.6 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Heavy chain, ... | | Authors: | Kumar, S, Sarkar, A, Wilson, I.A. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.602 Å) | | Cite: | HIV-1 vaccine design through minimizing envelope metastability.
Sci Adv, 4, 2018
|
|
7NYJ
 
 | | Structure of OBP1 from Varroa destructor, form P3<2>21 | | Descriptor: | CALCIUM ION, Odorant Binding Protein 1 from Varoa destructor, form P3<2>21, ... | | Authors: | Cambillau, C, Amigues, B, Roussel, A, Leone, P, Gaubert, A, Pelosi, P. | | Deposit date: | 2021-03-22 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A new non-classical fold of varroa odorant-binding proteins reveals a wide open internal cavity.
Sci Rep, 11, 2021
|
|
7NZA
 
 | | Structure of OBP1 from Varroa destructor, form P2<1> | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Odorant Binding Protein from Varroa destructor, form P2<1>, ... | | Authors: | Cambillau, C, Amigues, B, Roussel, A, Leone, P, Gaubert, A, Pelosi, P. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | A new non-classical fold of varroa odorant-binding proteins reveals a wide open internal cavity.
Sci Rep, 11, 2021
|
|
7FE5
 
 | |
4I3R
 
 | | Crystal structure of the outer domain of HIV-1 gp120 in complex with VRC-PG04 space group P3221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of VRC-PG04 Fab, Light chain of VRC-PG04 Fab, ... | | Authors: | Joyce, M.G, Biertumpfel, C, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2012-11-26 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Outer Domain of HIV-1 gp120: Antigenic Optimization, Structural Malleability, and Crystal Structure with Antibody VRC-PG04.
J.Virol., 87, 2013
|
|
