7V9I
 
 | | The Monomer mutant of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, DNA (5'-D(*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*CP*CP*AP*CP*GP*CP*GP*GP*T)-3') | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9H
 
 | | The BEN3 domain of protein Bend3 | | Descriptor: | BEN domain-containing protein 3 | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
7V9F
 
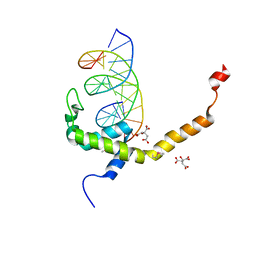 | | Selenomethionine mutant (L740Sem) of BEN4 domain of protein Bend3 with DNA | | Descriptor: | BEN domain-containing protein 3, CITRIC ACID, DNA (5'-D(*GP*CP*AP*CP*CP*GP*CP*GP*TP*GP*GP*GP*GP*CP*CP*A)-3'), ... | | Authors: | Zhang, J, Zhang, Y, You, Q, Huang, C, Zhang, T, Wang, M, Zhang, T, Yang, X, Xiong, J, Li, Y, Liu, C.P, Zhang, Z, Xu, R.M, Zhu, B. | | Deposit date: | 2021-08-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Highly enriched BEND3 prevents the premature activation of bivalent genes during differentiation.
Science, 375, 2022
|
|
4O2W
 
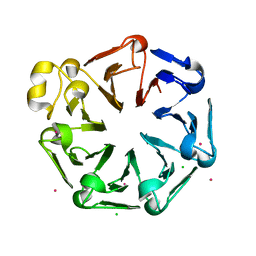 | | Crystal structure of the third RCC1-like domain of HERC1 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase HERC1, MAGNESIUM ION, ... | | Authors: | Dong, A, Hu, J, Li, Y, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the third RCC1-like domain of HERC1
To be Published
|
|
1RW9
 
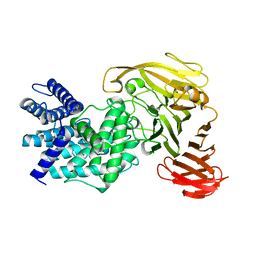 | | Crystal structure of the Arthrobacter aurescens chondroitin AC lyase | | Descriptor: | PHOSPHATE ION, SODIUM ION, chondroitin AC lyase | | Authors: | Lunin, V.V, Li, Y, Linhardt, R.J, Miyazono, H, Kyogashima, M, Kaneko, T, Bell, A.W, Cygler, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1P9N
 
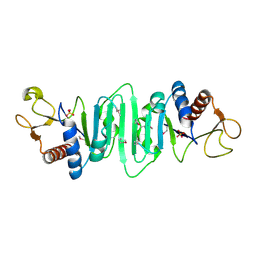 | | Crystal structure of Escherichia coli MobB. | | Descriptor: | Molybdopterin-guanine dinucleotide biosynthesis protein B, SULFATE ION | | Authors: | Rangarajan, S.E, Tocilj, A, Li, Y, Iannuzzi, P, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-05-12 | | Release date: | 2003-05-20 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecules of Escherichia coli MobB assemble into densely packed hollow cylinders in a crystal lattice with 75% solvent content.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1RWF
 
 | | Crystal structure of Arthrobacter aurescens chondroitin AC lyase in complex with chondroitin tetrasaccharide | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-2,6-anhydro-3-deoxy-L-xylo-hexonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Lunin, V.V, Li, Y, Miyazono, H, Kyogashima, M, Bell, A.W, Cygler, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1PS7
 
 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M. | | Deposit date: | 2003-06-20 | | Release date: | 2003-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
1Q18
 
 | | Crystal structure of E.coli glucokinase (Glk) | | Descriptor: | Glucokinase | | Authors: | Lunin, V.V, Li, Y, Schrag, J.D, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-07-18 | | Release date: | 2004-07-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structures of Escherichia coli ATP-dependent glucokinase and its complex with glucose.
J.Bacteriol., 186, 2004
|
|
4MVT
 
 | | Crystal structure of SUMO E3 Ligase PIAS3 | | Descriptor: | CHLORIDE ION, E3 SUMO-protein ligase PIAS3, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Hu, J, Li, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-09-24 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of SUMO E3 Ligase PIAS3
to be published
|
|
1RWG
 
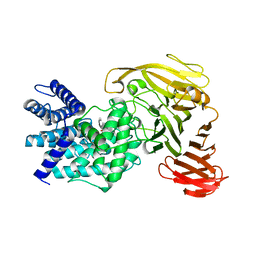 | | Crystal structure of Arthrobacter aurescens chondroitin AC lyase in complex with chondroitin tetrasaccharide | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-2,6-anhydro-3-deoxy-L-xylo-hexonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Lunin, V.V, Li, Y, Miyazono, H, Kyogashima, M, Bell, A.W, Cygler, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1RWH
 
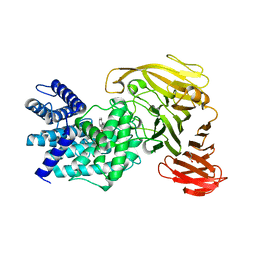 | | Crystal structure of Arthrobacter aurescens chondroitin AC lyase in complex with chondroitin tetrasaccharide | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-2,6-anhydro-3-deoxy-L-xylo-hexonic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lunin, V.V, Li, Y, Miyazono, H, Kyogashima, M, Bell, A.W, Cygler, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1PS6
 
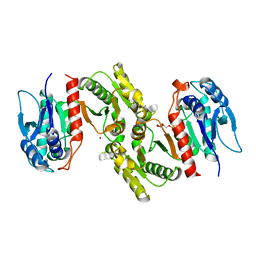 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-HYDROXY-L-THREONINE-5-MONOPHOSPHATE, 4-hydroxythreonine-4-phosphate dehydrogenase, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M. | | Deposit date: | 2003-06-20 | | Release date: | 2003-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
1RWA
 
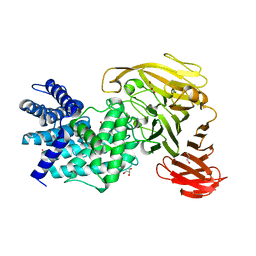 | | Crystal structure of Arthrobacter aurescens chondroitin AC lyase | | Descriptor: | GLYCEROL, MERCURY (II) ION, chondroitin AC lyase | | Authors: | Lunin, V.V, Li, Y, Miyazono, H, Kyogashima, M, Bell, A.W, Cygler, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1RWC
 
 | | Crystal structure of Arthrobacter aurescens chondroitin AC lyase | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lunin, V.V, Li, Y, Miyazono, H, Kyogashima, M, Bell, A.W, Cygler, M. | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme-substrate complex defines the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
6NBS
 
 | | WT ERK2 with compound 2507-8 | | Descriptor: | (5S)-5-benzyl-4,5-dihydro-1H-imidazol-2-amine, GLYCEROL, Mitogen-activated protein kinase 1, ... | | Authors: | Sammons, R.M, Perry, N.A, Cho, E.J, Kaoud, T.S, Zamora-Olivares, D.P, Piserchio, A, Houghten, R.A, Giulianotti, M, Li, Y, Debevec, G, Gurevich, V.V, Ghose, R, Iverson, T.M, Dalby, K.N. | | Deposit date: | 2018-12-10 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Class of Common Docking Domain Inhibitors That Prevent ERK2 Activation and Substrate Phosphorylation.
Acs Chem.Biol., 14, 2019
|
|
9J6O
 
 | | Crystal Structure of bromodomain of human CBP in complex with the inhibitor CZL-046 | | Descriptor: | (3~{S},5~{S})-1-[3,4-bis(fluoranyl)phenyl]-5-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]-3-fluoranyl-pyrrolidin-2-one, CHLORIDE ION, CREB-binding protein | | Authors: | Zhang, Y, Zhang, C, Chen, Z, Yang, H, Huang, X, Li, Y, Xu, Y. | | Deposit date: | 2024-08-16 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery of CZL-046 with an ( S )-3-Fluoropyrrolidin-2-one Scaffold as a p300 Bromodomain Inhibitor for the Treatment of Multiple Myeloma.
J.Med.Chem., 2024
|
|
8I2N
 
 | | The RIPK1 kinase domain in complex with QY7-2B compound | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 1, ~{N}-methyl-1-[4-[[[1-methyl-5-(phenylmethyl)pyrazol-3-yl]carbonylamino]methyl]phenyl]benzimidazole-5-carboxamide | | Authors: | Gong, X.Y, Li, Y, Meng, H.Y, Pan, L.F. | | Deposit date: | 2023-01-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-based development of potent and selective type-II kinase inhibitors of RIPK1.
Acta Pharm Sin B, 14, 2024
|
|
6OEB
 
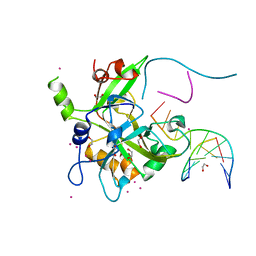 | | Crystal structure of HMCES SRAP domain in complex with 3' overhang DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*GP*AP*CP*GP*TP*T)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*G)-3'), ... | | Authors: | Halabelian, L, Ravichandran, M, Li, Y, Zeng, H, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3RCO
 
 | | Crystal structure of a conserved motif in human TDRD7 | | Descriptor: | CHLORIDE ION, Tudor domain-containing protein 7 | | Authors: | Dong, A, Xu, C, Walker, J.R, Lam, R, Guo, Y, Bian, C, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-03-31 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a conserved motif in human TDRD7
To be Published
|
|
6OEA
 
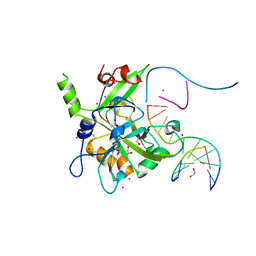 | | Crystal structure of HMCES SRAP domain in complex with longer 3' overhang DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*GP*AP*CP*GP*TP*TP*GP*TP*T)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*G)-3'), ... | | Authors: | Halabelian, L, Ravichandran, M, Li, Y, Zeng, H, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OOV
 
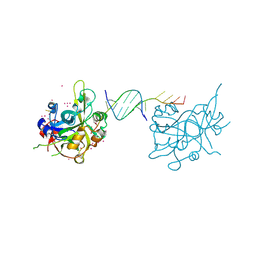 | | Crystal structure of HMCES SRAP domain in complex with palindromic 3' overhang DNA | | Descriptor: | DNA (5'-D(*CP*AP*AP*CP*GP*TP*TP*GP*TP*TP*TP*TP*T)-3'), Embryonic stem cell-specific 5-hydroxymethylcytosine-binding protein, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Zeng, H, Li, Y, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-23 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of HMCES SRAP domain in complex with palindromic 3' overhang DNA
To Be Published
|
|
6OE7
 
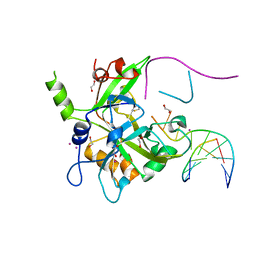 | | Crystal structure of HMCES cross-linked to DNA abasic site | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*GP*AP*CP*GP*TP*(DRZ)P*GP*TP*T)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*G)-3'), ... | | Authors: | Halabelian, L, Li, Y, Zeng, H, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6P0Q
 
 | | Crystal Structure of Ubiquitin-like Domain of Human WDR12 | | Descriptor: | 1,2-ETHANEDIOL, Ribosome biogenesis protein WDR12 | | Authors: | Halabelian, L, Dong, A, Zeng, H, Li, Y, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-17 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal Structure of Ubiquitin-like Domain of Human WDR12
to be published
|
|
3S4Y
 
 | | Crystal structure of human thiamin pyrophosphokinase 1 | | Descriptor: | CALCIUM ION, SULFATE ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Shen, L, Tempel, W, Tong, Y, Li, Y, Walker, J.R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human thiamin pyrophosphokinase 1
to be published
|
|
