4YG7
 
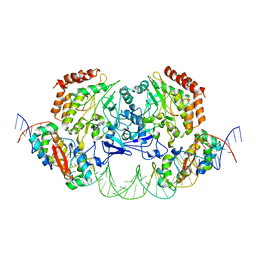 | | Structure of FL autorepression promoter complex | | Descriptor: | Antitoxin HipB, DNA (50-MER), Serine/threonine-protein kinase HipA | | Authors: | Schumacher, M.A. | | Deposit date: | 2015-02-25 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | HipBA-promoter structures reveal the basis of heritable multidrug tolerance.
Nature, 524, 2015
|
|
4ASK
 
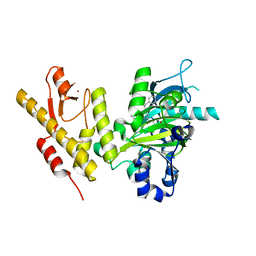 | | CRYSTAL STRUCTURE OF JMJD3 WITH GSK-J1 | | Descriptor: | 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, COBALT (II) ION, LYSINE-SPECIFIC DEMETHYLASE 6B, ... | | Authors: | Chung, C, Mosley, J, Liddle, J. | | Deposit date: | 2012-05-01 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A Selective Jumonji H3K27 Demethylase Inhibitor Modulates the Proinflammatory Macrophage Response
Nature, 488, 2012
|
|
1E77
 
 | |
4FC1
 
 | |
1LZN
 
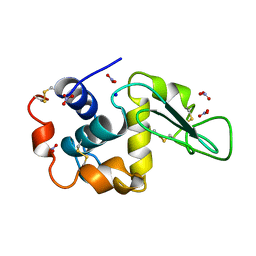 | | NEUTRON STRUCTURE OF HEN EGG-WHITE LYSOZYME | | Descriptor: | NITRATE ION, PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Bon, C.I, Lehmann, M.S, Wilkinson, C. | | Deposit date: | 1999-03-23 | | Release date: | 1999-04-01 | | Last modified: | 2023-12-27 | | Method: | NEUTRON DIFFRACTION (1.7 Å) | | Cite: | Quasi-Laue neutron-diffraction study of the water arrangement in crystals of triclinic hen egg-white lysozyme.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5AQ5
 
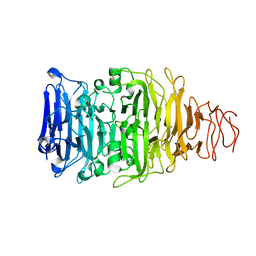 | |
5E5J
 
 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 6.0 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Kovalevsky, A.Y, Gerlits, O.O. | | Deposit date: | 2015-10-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5E5K
 
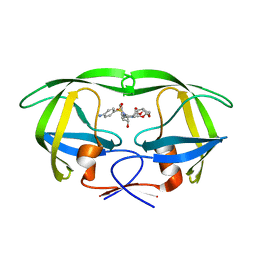 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 4.3 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 protease | | Authors: | Kovalevsky, A.Y, Das, A. | | Deposit date: | 2015-10-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.75 Å), X-RAY DIFFRACTION | | Cite: | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
4PWM
 
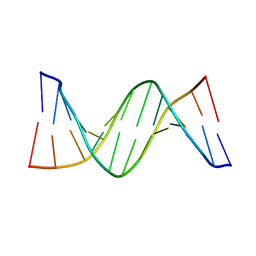 | | Crystal structure of Dickerson Drew Dodecamer with 5-carboxycytosine | | Descriptor: | 5'-[CGCGAATT(5CC)GCG]-3' | | Authors: | Szulik, M.W, Pallan, P, Banerjee, S, Voehler, M, Egli, M, Stone, M.P. | | Deposit date: | 2014-03-20 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Differential stabilities and sequence-dependent base pair opening dynamics of watson-crick base pairs with 5-hydroxymethylcytosine, 5-formylcytosine, or 5-carboxylcytosine.
Biochemistry, 54, 2015
|
|
4QC7
 
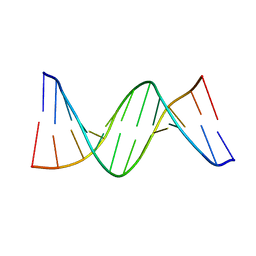 | | Dodecamer structure of 5-formylcytosine containing DNA | | Descriptor: | short DNA strands | | Authors: | Szulik, M.W, Pallan, P, Egli, M, Stone, M.P. | | Deposit date: | 2014-05-09 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential stabilities and sequence-dependent base pair opening dynamics of watson-crick base pairs with 5-hydroxymethylcytosine, 5-formylcytosine, or 5-carboxylcytosine.
Biochemistry, 54, 2015
|
|
4LWZ
 
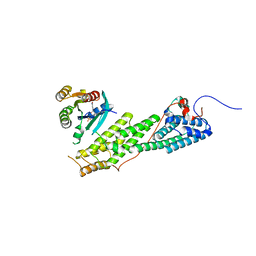 | | Crystal structure of Myo5b globular tail domain in complex with inactive Rab11a | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-11A, ... | | Authors: | Pylypenko, O, Attanda, W, Gauquelin, C, Houdusse, A. | | Deposit date: | 2013-07-29 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of myosin V Rab GTPase-dependent cargo recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4IAK
 
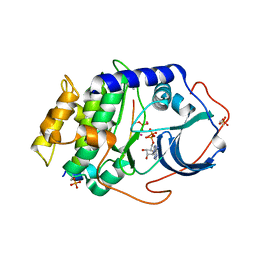 | |
4IAY
 
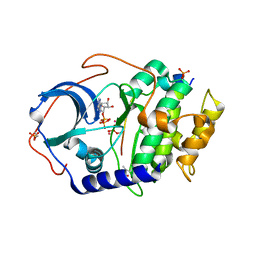 | |
4IAF
 
 | |
6EPK
 
 | | CRYSTAL STRUCTURE OF THE PRECURSOR MEMBRANE PROTEIN-ENVELOPE PROTEIN HETERODIMER FROM THE YELLOW FEVER VIRUS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, GLYCEROL, ... | | Authors: | Rey, F.A, Duquerroy, S, Crampon, E, Barba-Spaeth, G. | | Deposit date: | 2017-10-11 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | New insight into flavivirus maturation from structure/function studies of the yellow fever virus envelope protein complex
Mbio, 2023
|
|
4JFW
 
 | | Crystal structure of a bacterial fucosidase with iminosugar inhibitor (2S,3S,4R,5S)-2-[N-(propylferrocene)]aminoethyl-5-methylpyrrolidine-3,4-diol | | Descriptor: | (3alpha)-[3-({2-[(2S,3S,4R,5S)-3,4-dihydroxy-5-methylpyrrolidin-2-yl]ethyl}amino)propyl]ferrocene, IMIDAZOLE, SULFATE ION, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | alpha-L-fucosidase inhibition by pyrrolidine-ferrocene hybrids: rationalization of ligand-binding properties by structural studies.
Chemistry, 19, 2013
|
|
4JFV
 
 | | Crystal structure of a bacterial fucosidase with iminosugar inhibitor (2S,3S,4R,5S)-2-[N-(methylferrocene)]aminoethyl-5-methylpyrrolidine-3,4-diol | | Descriptor: | (3alpha)-[({2-[(2S,3S,4R,5S)-3,4-dihydroxy-5-methylpyrrolidin-2-yl]ethyl}amino)methyl]ferrocene, IMIDAZOLE, SULFATE ION, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | alpha-L-fucosidase inhibition by pyrrolidine-ferrocene hybrids: rationalization of ligand-binding properties by structural studies.
Chemistry, 19, 2013
|
|
4JFT
 
 | | Crystal structure of a bacterial fucosidase with iminosugar inhibitor N-desmethyl-4-epi-(+)-Codonopsinine | | Descriptor: | (2S,3S,4R,5S)-2-(4-methoxyphenyl)-5-methylpyrrolidine-3,4-diol, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | alpha-L-fucosidase inhibition by pyrrolidine-ferrocene hybrids: rationalization of ligand-binding properties by structural studies.
Chemistry, 19, 2013
|
|
4JFS
 
 | | Crystal structure of a bacterial fucosidase with iminosugar inhibitor 4-epi-(+)-Codonopsinine | | Descriptor: | (2S,3S,4R,5S)-2-(4-methoxyphenyl)-1,5-dimethylpyrrolidine-3,4-diol, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | alpha-L-fucosidase inhibition by pyrrolidine-ferrocene hybrids: rationalization of ligand-binding properties by structural studies.
Chemistry, 19, 2013
|
|
4IB0
 
 | |
4IAD
 
 | |
4IAZ
 
 | |
2YK0
 
 | | Structure of the N-terminal NTS-DBL1-alpha and CIDR-gamma double domain of the PfEMP1 protein from Plasmodium falciparum varO strain. | | Descriptor: | ERYTHROCYTE MEMBRANE PROTEIN 1, MAGNESIUM ION, SULFATE ION | | Authors: | Lewit-Bentley, A, Juillerat, A, Vigan-Womas, I, Guillotte, M, Hessel, A, Raynal, B, Mercereau-Puijalon, O, Bentley, G.A. | | Deposit date: | 2011-05-25 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Abo Blood-Group Dependence of Plasmodium Falciparum Rosetting.
Plos Pathog., 8, 2012
|
|
4PSY
 
 | | 100K crystal structure of Escherichia coli dihydrofolate reductase | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Wilson, M.A, Wan, Q, Bennet, B.C, Dealwis, C, Ringe, D, Petsko, G.A. | | Deposit date: | 2014-03-08 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Toward resolving the catalytic mechanism of dihydrofolate reductase using neutron and ultrahigh-resolution X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 22, 2014
|
|
4JFU
 
 | | Crystal structure of a bacterial fucosidase with iminosugar inhibitor | | Descriptor: | (2S,3R,4S,5S)-2-methyl-5-(4-methylphenyl)pyrrolidine-3,4-diol, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | alpha-L-fucosidase inhibition by pyrrolidine-ferrocene hybrids: rationalization of ligand-binding properties by structural studies.
Chemistry, 19, 2013
|
|
