4HKL
 
 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II Complexed with substrate (1.15 A) and Products (1.6 A) | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HKO
 
 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II (E177Q) in the apo form | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HKW
 
 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II Complexed with Substrate and Products | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Endo-1,4-beta-xylanase 2, ... | | Authors: | Kovalevsky, A.Y, Wan, Q, Langan, P, Coates, L. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HK8
 
 | | Crystal Structures of Mutant Endo- -1,4-xylanase II Complexed with substrate (1.15 A) and Products (1.6 A) | | Descriptor: | CITRIC ACID, Endo-1,4-beta-xylanase 2, GLYCEROL, ... | | Authors: | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.151 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4DUO
 
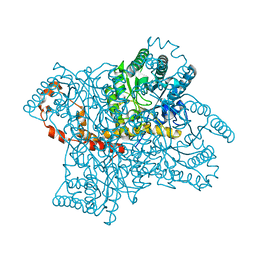 | | Room-temperature X-ray structure of D-Xylose Isomerase in complex with 2Mg2+ ions and xylitol at pH 7.7 | | Descriptor: | MAGNESIUM ION, Xylitol, Xylose isomerase | | Authors: | Kovalevsky, A.Y, Hanson, L, Langan, P. | | Deposit date: | 2012-02-22 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of D-xylose isomerase by polyols: atomic details by joint X-ray/neutron crystallography.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5B2Q
 
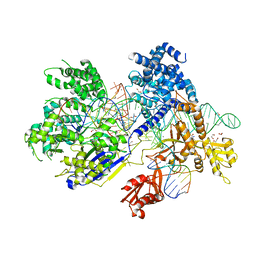 | | Crystal structure of Francisella novicida Cas9 RHA in complex with sgRNA and target DNA (TGG PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Hirano, H, Nishimasu, H, Nakane, T, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-01 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Engineering of Francisella novicida Cas9
Cell, 164, 2016
|
|
6WYH
 
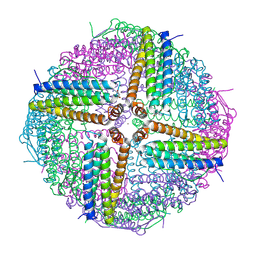 | |
9ARD
 
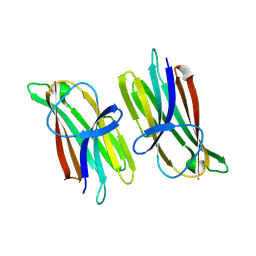 | |
6WYG
 
 | |
6WYF
 
 | |
5B2P
 
 | | Crystal structure of Francisella novicida Cas9 in complex with sgRNA and target DNA (TGA PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Hirano, H, Nishimasu, H, Nakane, T, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-01 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Engineering of Francisella novicida Cas9
Cell, 164, 2016
|
|
6HQ0
 
 | | Crystal structure of ENL (MLLT1), apo form | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
6WPM
 
 | | Crystal structure of a putative oligosaccharide periplasmic-binding protein from Synechococcus sp. MITs9220 in complex with zinc | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Ford, B.A, Michie, K.A, Paulsen, I.T, Mabbutt, B.C, Shah, B.S. | | Deposit date: | 2020-04-27 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Novel functional insights into a modified sugar-binding protein from Synechococcus MITS9220.
Sci Rep, 12, 2022
|
|
6HPY
 
 | | Crystal structure of ENL (MLLT1) in complex with compound 12 | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-[(4-~{tert}-butylphenyl)carbonylamino]phenyl]propanoic acid, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
2DVW
 
 | |
7LM4
 
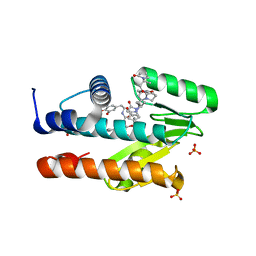 | | The crystal structure of the I38T mutant PA Endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988503 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-2-(2-methylphenyl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, Rankovic, Z, White, S.W. | | Deposit date: | 2021-02-05 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6XR3
 
 | |
6HPX
 
 | | Crystal structure of ENL (MLLT1) in complex with compound 19 | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL, ~{N}-[(3-chlorophenyl)methyl]-1-(2-pyrrolidin-1-ylethyl)benzimidazole-5-carboxamide | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
9B9P
 
 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase in complex with Mg2+, and JK-5-114 | | Descriptor: | 6-hydroxy-2-phenyl[1,3]thiazolo[4,5-d]pyrimidine-5,7(4H,6H)-dione, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION | | Authors: | Kurz, J.L, Lin, X, Guddat, L.W. | | Deposit date: | 2024-04-03 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | A ketol-acid reductoisomerase inhibitor that has antituberculosis and herbicidal activity.
Chemistry, 2025
|
|
9CE6
 
 | |
7ZGE
 
 | |
6HPZ
 
 | | Crystal structure of ENL (MLLT1) in complex with acetyllysine | | Descriptor: | 1,2-ETHANEDIOL, N(6)-ACETYLLYSINE, Protein ENL | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
5B2O
 
 | | Crystal structure of Francisella novicida Cas9 in complex with sgRNA and target DNA (TGG PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Hirano, H, Nishimasu, H, Nakane, T, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-01 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Engineering of Francisella novicida Cas9
Cell, 164, 2016
|
|
8V1X
 
 | | Crystal structure of polyketide synthase (PKS) thioreductase domain from Streptomyces coelicolor | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Pereira, J.H, Dan, Q, Keasling, J, Adams, P.D. | | Deposit date: | 2023-11-21 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Polyketide synthase-based biosynthesis of diols and aldehyde derivatives via terminal thioreduction
To Be Published
|
|
4HK9
 
 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II Complexed with substrate (1.15 A) and Products (1.6 A) | | Descriptor: | Endo-1,4-beta-xylanase 2, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
