1GTP
 
 | | GTP CYCLOHYDROLASE I | | Descriptor: | GTP CYCLOHYDROLASE I, SULFATE ION | | Authors: | Nar, H, Huber, R, Meining, W, Bacher, A. | | Deposit date: | 1995-09-16 | | Release date: | 1996-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Atomic structure of GTP cyclohydrolase I.
Structure, 3, 1995
|
|
1GTL
 
 | | The thermostable serine-carboxyl type proteinase, kumamolisin (KSCP) - complex with Ac-Ile-Pro-Phe-cho | | Descriptor: | ALDEHYDE INHIBITOR, CALCIUM ION, KUMAMOLYSIN, ... | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-16 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1H98
 
 | | New Insights into Thermostability of Bacterial Ferredoxins: High Resolution Crystal Structure of the Seven-Iron Ferredoxin from Thermus thermophilus | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Macedo-Ribeiro, S, Martins, B.M, Pereira, P.J.B, Buse, G, Huber, R, Soulimane, T. | | Deposit date: | 2001-03-05 | | Release date: | 2001-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | New Insights Into the Thermostability of Bacterial Ferredoxins: High-Resolution Crystal Structure of the Seven-Iron Ferredoxin from Thermus Thermophilus
J.Biol.Inorg.Chem., 6, 2001
|
|
1GTJ
 
 | | Crystal structure of the thermostable serine-carboxyl type proteinase, kumamolisin (KSCP) - complex with Ac-Ile-Ala-Phe-cho | | Descriptor: | ALDEHYDE INHIBITOR, CALCIUM ION, KUMAMOLYSIN, ... | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-15 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1H59
 
 | | Complex of IGFBP-5 with IGF-I | | Descriptor: | INSULIN-LIKE GROWTH FACTOR BINDING PROTEIN 5, INSULIN-LIKE GROWTH FACTOR IA | | Authors: | Zeslawski, W, Beisel, H.G, Kamionka, M, Kalus, W, Engh, R.A, Huber, R, Holak, T.A. | | Deposit date: | 2001-05-21 | | Release date: | 2002-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Interaction of Insulin-Like Growth Factor-I with the N-Terminal Domain of Igfbp-5
Embo J., 20, 2001
|
|
1GV3
 
 | | The 2.0 Angstrom resolution structure of the catalytic portion of a cyanobacterial membrane-bound manganese superoxide dismutase | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Atzenhofer, W, Regelsberger, G, Jacob, U, Huber, R, Peschek, G.A, Obinger, C. | | Deposit date: | 2002-02-05 | | Release date: | 2002-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0A Resolution Structure of the Catalytic Portion of a Cyanobacterial Membrane-Bound Manganese Superoxide Dismutase
J.Mol.Biol., 321, 2002
|
|
1GT9
 
 | | High resolution crystal structure of a thermostable serine-carboxyl type proteinase, kumamolisin (kscp) | | Descriptor: | CALCIUM ION, KUMAMOLYSIN, SULFATE ION | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-14 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1HOE
 
 | |
1HLE
 
 | | CRYSTAL STRUCTURE OF CLEAVED EQUINE LEUCOCYTE ELASTASE INHIBITOR DETERMINED AT 1.95 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, HORSE LEUKOCYTE ELASTASE INHIBITOR | | Authors: | Baumann, U, Bode, W, Huber, R, Travis, J, Potempa, J. | | Deposit date: | 1992-04-13 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of cleaved equine leucocyte elastase inhibitor determined at 1.95 A resolution.
J.Mol.Biol., 226, 1992
|
|
2FCB
 
 | | HUMAN FC GAMMA RECEPTOR IIB ECTODOMAIN (CD32) | | Descriptor: | PROTEIN (FC GAMMA RIIB) | | Authors: | Sondermann, P, Huber, R, Jacob, U. | | Deposit date: | 1999-01-07 | | Release date: | 2000-03-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of the soluble form of the human fcgamma-receptor IIb: a new member of the immunoglobulin superfamily at 1.7 A resolution.
EMBO J., 18, 1999
|
|
1JFD
 
 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE | | Descriptor: | INORGANIC PYROPHOSPHATASE, SULFATE ION | | Authors: | Oganesyan, V, Avaeva, S.M, Huber, R, Harutyunyan, E.H. | | Deposit date: | 1997-05-31 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Escherichia coli inorganic pyrophosphatase complexed with SO4(2-). Ligand-induced molecular asymmetry.
FEBS Lett., 410, 1997
|
|
2DSP
 
 | | Structural Basis for the Inhibition of Insulin-like Growth Factors by IGF Binding Proteins | | Descriptor: | Insulin-like growth factor IB, Insulin-like growth factor-binding protein 4 | | Authors: | Sitar, T, Popowicz, G.M, Siwanowicz, I, Huber, R, Holak, T.A. | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DSR
 
 | | Structural Basis for the Inhibition of Insulin-like Growth Factors by IGF Binding Proteins | | Descriptor: | Insulin-like growth factor IB, Insulin-like growth factor-binding protein 4 | | Authors: | Sitar, T, Popowicz, G.M, Siwanowicz, I, Huber, R, Holak, T.A. | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1QGN
 
 | | CYSTATHIONINE GAMMA-SYNTHASE FROM NICOTIANA TABACUM | | Descriptor: | PROTEIN (CYSTATHIONINE GAMMA-SYNTHASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Steegborn, C, Messerschmidt, A, Laber, B, Streber, W, Huber, R, Clausen, T. | | Deposit date: | 1999-05-02 | | Release date: | 1999-08-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of cystathionine gamma-synthase from Nicotiana tabacum reveals its substrate and reaction specificity.
J.Mol.Biol., 290, 1999
|
|
1Q8U
 
 | | The Catalytic Subunit of cAMP-dependent Protein Kinase in Complex with Rho-kinase Inhibitor H-1152P | | Descriptor: | (S)-2-METHYL-1-[(4-METHYL-5-ISOQUINOLINE)SULFONYL]-HOMOPIPERAZINE, N-OCTANOYL-N-METHYLGLUCAMINE, cAMP-dependent protein kinase inhibitor, ... | | Authors: | Breitenlechner, C, Gassel, M, Hidaka, H, Kinzel, V, Huber, R, Engh, R.A, Bossemeyer, D. | | Deposit date: | 2003-08-22 | | Release date: | 2003-12-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein kinase A in complex with Rho-kinase inhibitors Y-27632, Fasudil, and H-1152P: structural basis of selectivity.
Structure, 11, 2003
|
|
1Q8W
 
 | | The Catalytic Subunit of cAMP-dependent Protein Kinase in Complex with Rho-kinase Inhibitor Fasudil (HA-1077) | | Descriptor: | 5-(1,4-DIAZEPAN-1-SULFONYL)ISOQUINOLINE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C, Gassel, M, Hidaka, H, Kinzel, V, Huber, R, Engh, R.A, Bossemeyer, D. | | Deposit date: | 2003-08-22 | | Release date: | 2003-12-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein kinase A in complex with Rho-kinase inhibitors Y-27632, Fasudil, and H-1152P: structural basis of selectivity.
Structure, 11, 2003
|
|
1PO9
 
 | | Crytsal structure of isoaspartyl dipeptidase | | Descriptor: | Isoaspartyl dipeptidase, ZINC ION | | Authors: | Jozic, D, Kaiser, J.T, Huber, R, Bode, W, Maskos, K. | | Deposit date: | 2003-06-15 | | Release date: | 2004-06-22 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of isoaspartyl dipeptidase from E.coli: a dinuclear zinc peptidase evolved from amidohydrolases.
J.Mol.Biol., 332, 2003
|
|
1RFN
 
 | | HUMAN COAGULATION FACTOR IXA IN COMPLEX WITH P-AMINO BENZAMIDINE | | Descriptor: | CALCIUM ION, P-AMINO BENZAMIDINE, PROTEIN (COAGULATION FACTOR IX), ... | | Authors: | Hopfner, K.-P, Lang, A, Karcher, A, Sichler, K, Kopetzki, E, Brandstetter, H, Huber, R, Bode, W, Engh, R.A. | | Deposit date: | 1999-04-19 | | Release date: | 1999-09-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Coagulation factor IXa: the relaxed conformation of Tyr99 blocks substrate binding.
Structure Fold.Des., 7, 1999
|
|
6GTH
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Wiedorn, M, Oberthuer, D, Werner, N, Schubert, R, White, T.A, Mancuso, A, Perbandt, M, Betzel, C, Barty, A, Chapman, H. | | Deposit date: | 2018-06-18 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
1Q8T
 
 | | The Catalytic Subunit of cAMP-dependent Protein Kinase (PKA) in Complex with Rho-kinase Inhibitor Y-27632 | | Descriptor: | (R)-TRANS-4-(1-AMINOETHYL)-N-(4-PYRIDYL) CYCLOHEXANECARBOXAMIDE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C, Gassel, M, Hidaka, H, Kinzel, V, Huber, R, Engh, R.A, Bossemeyer, D. | | Deposit date: | 2003-08-22 | | Release date: | 2003-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein kinase A in complex with Rho-kinase inhibitors Y-27632, Fasudil, and H-1152P: structural basis of selectivity.
Structure, 11, 2003
|
|
1SMH
 
 | | Protein kinase A variant complex with completely ordered N-terminal helix | | Descriptor: | (R,R)-2,3-BUTANEDIOL, N-OCTANOYL-N-METHYLGLUCAMINE, cAMP-Dependent Protein Kinase, ... | | Authors: | Breitenlechner, C, Engh, R.A, Huber, R, Kinzel, V, Bossemeyer, D, Gassel, M. | | Deposit date: | 2004-03-09 | | Release date: | 2004-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.044 Å) | | Cite: | The Typically Disordered N-Terminus of PKA Can Fold as a Helix and Project the Myristoylation Site into Solution
Biochemistry, 43, 2004
|
|
1SP8
 
 | | 4-Hydroxyphenylpyruvate Dioxygenase | | Descriptor: | 4-Hydroxyphenylpyruvate Dioxygenase, FE (II) ION | | Authors: | Fritze, I.M, Linden, L, Freigang, J, Auerbach, G, Huber, R, Steinbacher, S. | | Deposit date: | 2004-03-16 | | Release date: | 2004-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structures of Zea mays and Arabidopsis 4-Hydroxyphenylpyruvate Dioxygenase
Plant physiol., 134, 2004
|
|
1SP9
 
 | | 4-Hydroxyphenylpyruvate Dioxygenase | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, FE (II) ION | | Authors: | Fritze, I.M, Linden, L, Freigang, J, Auerbach, G, Huber, R, Steinbacher, S. | | Deposit date: | 2004-03-16 | | Release date: | 2004-09-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structures of Zea mays and Arabidopsis 4-Hydroxyphenylpyruvate Dioxygenase
Plant Physiol., 134, 2004
|
|
1SQL
 
 | | Crystal structure of 7,8-dihydroneopterin aldolase in complex with guanine | | Descriptor: | GUANINE, dihydroneopterin aldolase | | Authors: | Bauer, S, Schott, A.K, Illarionova, V, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2004-03-19 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biosynthesis of Tetrahydrofolate in Plants: Crystal Structure of 7,8-Dihydroneopterin Aldolase from Arabidopsis thaliana Reveals a Novel Adolase Class.
J.Mol.Biol., 339, 2004
|
|
1RYP
 
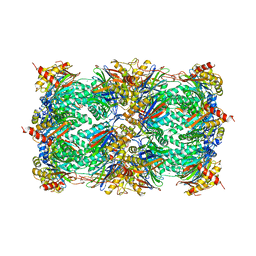 | | CRYSTAL STRUCTURE OF THE 20S PROTEASOME FROM YEAST AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | 20S PROTEASOME, MAGNESIUM ION | | Authors: | Groll, M, Ditzel, L, Loewe, J, Stock, D, Bochtler, M, Bartunik, H.D, Huber, R. | | Deposit date: | 1997-02-26 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of 20S proteasome from yeast at 2.4 A resolution.
Nature, 386, 1997
|
|
