3HVN
 
 | | Crystal structure of cytotoxin protein suilysin from Streptococcus suis | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, HEPTANE-1,2,3-TRIOL, Hemolysin | | Authors: | Xu, L, Huang, B, Du, H, Zhang, C.X, Xu, J, Li, X, Rao, Z. | | Deposit date: | 2009-06-16 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.852 Å) | | Cite: | Crystal structure of cytotoxin protein suilysin from Streptococcus suis.
Protein Cell, 1, 2010
|
|
2OQI
 
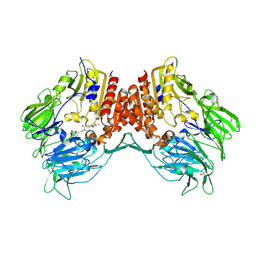 | | Human Dipeptidyl Peptidase IV (DPP4) with Piperidinone-constrained phenethylamine | | Descriptor: | (4R,5R)-5-AMINO-1-[2-(1,3-BENZODIOXOL-5-YL)ETHYL]-4-(2,4,5-TRIFLUOROPHENYL)PIPERIDIN-2-ONE, Dipeptidyl peptidase 4 (Dipeptidyl peptidase IV) (DPP IV) (T-cell activation antigen CD26) (TP103) (Adenosine deaminase complexing protein 2) (ADABP) | | Authors: | Pei, Z, Li, X, von Geldern, T.W, Longenecker, K.L, Pireh, D, Stewart, K.D, Backes, B.J, Lai, C, Lubben, T.H, Ballaron, S.J, Beno, D.W, Kempf-Grote, A.J, Sham, H.L, Trevillyan, J.M. | | Deposit date: | 2007-01-31 | | Release date: | 2007-04-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Structure-Activity Relationships of Piperidinone- and Piperidine-Constrained Phenethylamines as Novel, Potent, and Selective Dipeptidyl Peptidase IV Inhibitors.
J.Med.Chem., 50, 2007
|
|
8WZ2
 
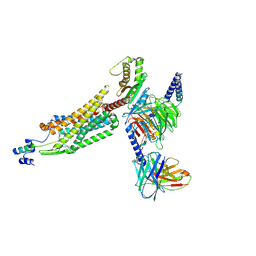 | | Structure of 26RFa-pyroglutamylated RFamide peptide receptor complex | | Descriptor: | G-alpha q, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jin, S, Li, X, Xu, Y, Guo, S, Wu, C, Zhang, H, Yuan, Q, Xu, H.E, Xie, X, Jiang, Y. | | Deposit date: | 2023-11-01 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis for recognition of 26RFa by the pyroglutamylated RFamide peptide receptor.
Cell Discov, 10, 2024
|
|
4KDR
 
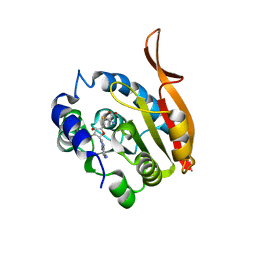 | | Crystal Structure of UBIG/SAH complex | | Descriptor: | 3-demethylubiquinone-9 3-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zhu, Y, Teng, M, Li, X. | | Deposit date: | 2013-04-25 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Crystal Structure of the UBIG/SAH complex
To be Published
|
|
8TWU
 
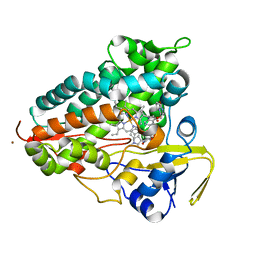 | | Crystal structure of Cytochrome P450 AspB bound to N1-methylated cyclo-L-Trp-L-Pro | | Descriptor: | (3S,5S,8aS)-3-[(1-methyl-1H-indol-3-yl)methyl]hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, Cytochrome P450 AspB, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gering, H.E, Li, X, Tang, H, Swartz, P.D, Chang, W.-C, Makris, T.M. | | Deposit date: | 2023-08-21 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A Ferric-Superoxide Intermediate Initiates P450-Catalyzed Cyclic Dipeptide Dimerization.
J.Am.Chem.Soc., 145, 2023
|
|
2OQV
 
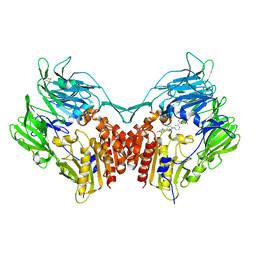 | | Human Dipeptidyl Peptidase IV (DPP4) with piperidine-constrained phenethylamine | | Descriptor: | (3R,4R)-1-{6-[3-(METHYLSULFONYL)PHENYL]PYRIMIDIN-4-YL}-4-(2,4,5-TRIFLUOROPHENYL)PIPERIDIN-3-AMINE, Dipeptidyl peptidase 4 (Dipeptidyl peptidase IV) (DPP IV) | | Authors: | Pei, Z, Li, X, von Geldern, T.W, Longenecker, K.L, Pireh, D, Stewart, K.D. | | Deposit date: | 2007-02-01 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Structure-Activity Relationships of Piperidinone- and Piperidine-Constrained Phenethylamines as Novel, Potent, and Selective Dipeptidyl Peptidase IV Inhibitors.
J.Med.Chem., 50, 2007
|
|
1C9W
 
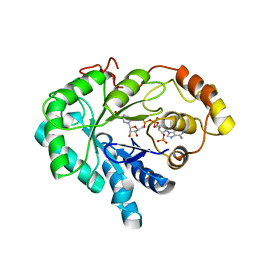 | | CHO REDUCTASE WITH NADP+ | | Descriptor: | CHO REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ye, Q, Li, X, Hyndman, D, Flynn, T.G, Jia, Z. | | Deposit date: | 1999-08-03 | | Release date: | 2000-01-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of CHO reductase, a member of the aldo-keto reductase superfamily.
Proteins, 38, 2000
|
|
3TO8
 
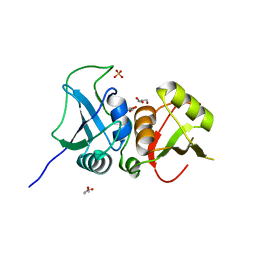 | | Crystal structure of the two C-terminal RRM domains of heterogeneous nuclear ribonucleoprotein L (hnRNP L) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Heterogeneous nuclear ribonucleoprotein L, ... | | Authors: | Zhang, W.J, Zeng, F.X, Liu, Y.W, Zhao, Y, Niu, L.W, Teng, M.K, Li, X. | | Deposit date: | 2011-09-04 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of the two C-terminal RRM domains of heterogeneous nuclear ribonucleoprotein L (hnRNP L)
To be Published
|
|
3KXL
 
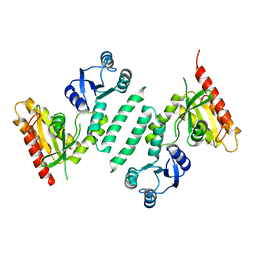 | | crystal structure of SsGBP mutation variant G235S | | Descriptor: | GTP-binding protein (HflX), THIOCYANATE ION | | Authors: | Huang, B, Li, X, Zhang, X.C, Rao, Z. | | Deposit date: | 2009-12-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional study on GTP hydrolysis by the GTP binding protein from Sulfolobus solfataricus, a member of the HflX family.
J.Biochem., 2010
|
|
3KXI
 
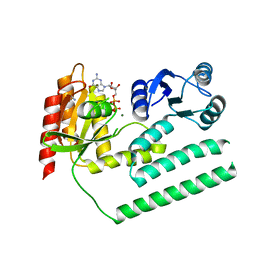 | | crystal structure of SsGBP and GDP complex | | Descriptor: | GTP-binding protein (HflX), GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, B, Li, X, Zhang, X.C, Rao, Z. | | Deposit date: | 2009-12-03 | | Release date: | 2010-05-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Functional study on GTP hydrolysis by the GTP binding protein from Sulfolobus solfataricus, a member of the HflX family.
J.Biochem., 2010
|
|
3LBF
 
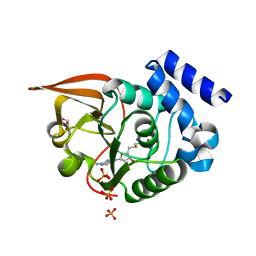 | | Crystal structure of Protein L-isoaspartyl methyltransferase from Escherichia coli | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein-L-isoaspartate O-methyltransferase, ... | | Authors: | Fang, P, Li, X, Wang, J, Niu, L, Teng, M. | | Deposit date: | 2010-01-08 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the protein L-isoaspartyl methyltransferase from Escherichia coli
Cell Biochem.Biophys., 58, 2010
|
|
3LKX
 
 | | Human nac dimerization domain | | Descriptor: | Nascent polypeptide-associated complex subunit alpha, Transcription factor BTF3 | | Authors: | Liu, Y, Hu, Y, Li, X, Niu, L, Teng, M. | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the human nascent polypeptide-associated complex domain reveals a nucleic acid-binding region on the NACA subunit
Biochemistry, 49, 2010
|
|
5AGT
 
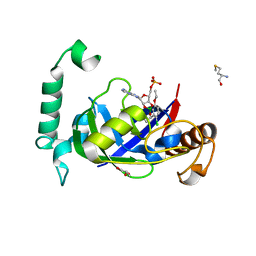 | | Crystal structure of the LeuRS editing domain of Mycobacterium tuberculosis in complex with the adduct (S)-3-(Aminomethyl)-4-chloro-7-ethoxybenzo[c][1,2]oxaborol-1(3H)-ol-AMP | | Descriptor: | 4-Chloro-3-aminomethyl-7-[ethoxy]-3H-benzo[C][1,2]oxaborol-1-ol modified adenosine, GLYCEROL, LEUCINE--TRNA LIGASE, ... | | Authors: | Palencia, A, Li, X, Alley, M.R.K, Ding, C, Easom, E.E, Hernandez, V, Meewan, M, Mohan, M, Rock, F.L, Franzblau, S.G, Wang, Y, Lenaerts, A.J, Parish, T, Cooper, C.B, Waters, M.G, Ma, Z, Mendoza, A, Barros, D, Cusack, S, Plattner, J.J. | | Deposit date: | 2015-02-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of Novel Oral Protein Synthesis Inhibitors of Mycobacterium Tuberculosis that Target Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5AGR
 
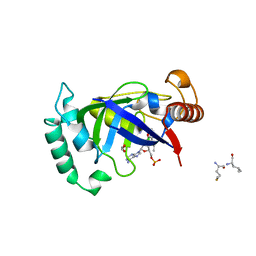 | | Crystal structure of the LeuRS editing domain of Mycobacterium tuberculosis in complex with the adduct (S)-3-(Aminomethyl)-7-ethoxybenzo[c][1,2]oxaborol-1(3H)-ol-AMP | | Descriptor: | 1,2-ETHANEDIOL, 3-AMINOMETHYL-7-(ETHOXY)-3H-BENZO[C][1,2]OXABOROL-1-OL modified adenosine, LEUCINE, ... | | Authors: | Palencia, A, Li, X, Alley, M.R.K, Ding, C, Easom, E.E, Hernandez, V, Meewan, M, Mohan, M, Rock, F.L, Franzblau, S.G, Wang, Y, Lenaerts, A.J, Parish, T, Cooper, C.B, Waters, M.G, Ma, Z, Mendoza, A, Barros, D, Cusack, S, Plattner, J.J. | | Deposit date: | 2015-02-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of Novel Oral Protein Synthesis Inhibitors of Mycobacterium Tuberculosis that Target Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
6LH8
 
 | | Structure of aerolysin-like protein (Bombina maxima) | | Descriptor: | aerolysin-like protein | | Authors: | Bian, X.L, Wang, Q.Q, Li, X, Teng, M.Q, Zhang, Y. | | Deposit date: | 2019-12-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | A cellular endolysosome-modulating pore-forming protein from a toad is negatively regulated by its paralog under oxidizing conditions.
J.Biol.Chem., 295, 2020
|
|
5AGS
 
 | | Crystal structure of the LeuRS editing domain of Mycobacterium tuberculosis in complex with the adduct 3-(Aminomethyl)-4-bromo-7-ethoxybenzo[c][1,2]oxaborol-1(3H)-ol-AMP | | Descriptor: | 3-(AMINOMETHYL)-4-BROMO-7-ETHOXYBENZO[C][1,2]OXABOROL-1(3H)-OL-MODIFIED ADENOSINE, LEUCYL-TRNA SYNTHETASE, METHIONINE | | Authors: | Palencia, A, Li, X, Alley, M.R.K, Ding, C, Easom, E.E, Hernandez, V, Meewan, M, Mohan, M, Rock, F.L, Franzblau, S.G, Wang, Y, Lenaerts, A.J, Parish, T, Cooper, C.B, Waters, M.G, Ma, Z, Mendoza, A, Barros, D, Cusack, S, Plattner, J.J. | | Deposit date: | 2015-02-03 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of Novel Oral Protein Synthesis Inhibitors of Mycobacterium Tuberculosis that Target Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
6LHZ
 
 | | Structure of aerolysin-like protein (Bombina maxima) | | Descriptor: | aerolysin-like protein | | Authors: | Bian, X.L, Wang, Q.Q, Li, X, Teng, M.Q, Zhang, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | A cellular endolysosome-modulating pore-forming protein from a toad is negatively regulated by its paralog under oxidizing conditions.
J.Biol.Chem., 295, 2020
|
|
3MNM
 
 | | Crystal structure of GAE domain of GGA2p from Saccharomyces cerevisiae | | Descriptor: | ADP-ribosylation factor-binding protein GGA2, GLYCEROL, SULFATE ION | | Authors: | Fang, P, Wang, J, Li, X, Niu, L, Teng, M. | | Deposit date: | 2010-04-21 | | Release date: | 2010-09-08 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural basis for the specificity of the GAE domain of yGGA2 for its accessory proteins Ent3 and Ent5
Biochemistry, 49, 2010
|
|
5CN2
 
 | |
5CMY
 
 | | Crystal structure of yeast Ent5 N-terminal domain-native | | Descriptor: | Epsin-5, GLYCEROL | | Authors: | Zhang, F, Song, Y, Li, X, Teng, M.K. | | Deposit date: | 2015-07-17 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and functional insight into the N-terminal domain of the clathrin adaptor Ent5 from Saccharomyces cerevisiae
Biochem.Biophys.Res.Commun., 477, 2016
|
|
5CMW
 
 | | Crystal structure of yeast Ent5 N-terminal domain-soaked in KI | | Descriptor: | Epsin-5, GLYCEROL, IODIDE ION | | Authors: | Zhang, F, Song, Y, Li, X, Teng, M.K. | | Deposit date: | 2015-07-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insight into the N-terminal domain of the clathrin adaptor Ent5 from Saccharomyces cerevisiae
Biochem.Biophys.Res.Commun., 477, 2016
|
|
7WLT
 
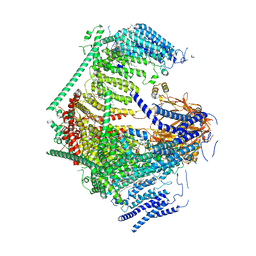 | | the Curved Structure of mPIEZO1 in Lipid Bilayer | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, ... | | Authors: | Yang, X, Lin, C, Chen, X, Li, S, Li, X, Xiao, B. | | Deposit date: | 2022-01-13 | | Release date: | 2022-04-13 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure deformation and curvature sensing of PIEZO1 in lipid membranes.
Nature, 604, 2022
|
|
7WLU
 
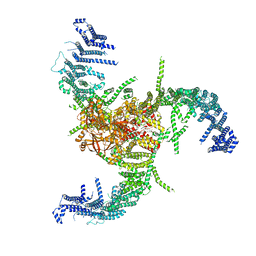 | | The Flattened Structure of mPIEZO1 in Lipid Bilayer | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, Piezo-type mechanosensitive ion channel component 1 | | Authors: | Yang, X, Lin, C, Chen, X, Li, S, Li, X, Xiao, B. | | Deposit date: | 2022-01-13 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (6.81 Å) | | Cite: | Structure deformation and curvature sensing of PIEZO1 in lipid membranes.
Nature, 604, 2022
|
|
3VBC
 
 | |
3KXK
 
 | |
