9CR2
 
 | | TRiC-ADP-C1 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2024-07-20 | | Release date: | 2025-01-22 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | The conformational landscape of TRiC ring-opening and its underlying stepwise mechanism revealed by cryo-EM.
Qrb Discov, 6, 2025
|
|
9CS4
 
 | | TRiC-ADP-S3 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2024-07-23 | | Release date: | 2025-01-22 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | The conformational landscape of TRiC ring-opening and its underlying stepwise mechanism revealed by cryo-EM.
Qrb Discov, 6, 2025
|
|
9CS6
 
 | | TRiC-ADP-S5 state is a conformation when TRiC incubated in 1 mM ADP | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2024-07-23 | | Release date: | 2025-01-22 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The conformational landscape of TRiC ring-opening and its underlying stepwise mechanism revealed by cryo-EM.
Qrb Discov, 6, 2025
|
|
8WVB
 
 | | Crystal structure of Lsd18 mutant S195M | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Putative epoxidase LasC | | Authors: | Liu, N, Xiao, H.L, Chen, X. | | Deposit date: | 2023-10-23 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Simultaneous Improvement in the Thermostability and Catalytic Activity of Epoxidase Lsd18 for the Synthesis of Lasalocid A.
Int J Mol Sci, 24, 2023
|
|
8WVF
 
 | | Crystal structure of Lsd18 mutant T189M and S195M | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative epoxidase LasC | | Authors: | Liu, N, Xiao, H.L, Chen, X. | | Deposit date: | 2023-10-23 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.757 Å) | | Cite: | Simultaneous Improvement in the Thermostability and Catalytic Activity of Epoxidase Lsd18 for the Synthesis of Lasalocid A.
Int J Mol Sci, 24, 2023
|
|
6VLU
 
 | | Factor XIa in complex with compound 7 | | Descriptor: | CITRIC ACID, Coagulation factor XIa light chain, N-cyclohexyl-D-leucyl-N-[(1-aminoisoquinolin-6-yl)methyl]-4,4-difluoro-L-prolinamide | | Authors: | Lesburg, C.A, Fradera, X. | | Deposit date: | 2020-01-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Augmenting Hit Identification by Virtual Screening Techniques in Small Molecule Drug Discovery.
J.Chem.Inf.Model., 60, 2020
|
|
6VLM
 
 | |
6VLV
 
 | | Factor XIa in complex with compound 11 | | Descriptor: | 1-carbamimidamido-4-chloro-N-[(2R)-3-methyl-1-(morpholin-4-yl)-1-oxobutan-2-yl]isoquinoline-7-sulfonamide, CITRATE ANION, Coagulation factor XIa light chain | | Authors: | Lesburg, C.A. | | Deposit date: | 2020-01-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Augmenting Hit Identification by Virtual Screening Techniques in Small Molecule Drug Discovery.
J.Chem.Inf.Model., 60, 2020
|
|
4O0U
 
 | |
4O0S
 
 | |
4O0W
 
 | |
8Y42
 
 | | Crystal structure of SARS-CoV-2 3CL protease (3CLpro) in complex with compound 51 | | Descriptor: | 3C-like proteinase nsp5, ~{N}-[(1~{S},2~{R})-2-[[4-cyclopropyl-2-(methylcarbamoyl)-6-nitro-phenyl]amino]cyclohexyl]-2-oxidanylidene-1~{H}-quinoline-4-carboxamide | | Authors: | Nie, T.Q, Su, H.X, Li, M.J, Xu, Y.C. | | Deposit date: | 2024-01-30 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Novel Nonpeptidic and Noncovalent Small Molecule 3CL pro Inhibitors as anti-SARS-CoV-2 Drug Candidate.
J.Med.Chem., 67, 2024
|
|
8Y44
 
 | | Crystal structure of SARS-CoV-2 3CL protease (3CLpro) in complex with compound 44 | | Descriptor: | 3C-like proteinase nsp5, ~{N}-[(1~{S},2~{R})-2-[(4-bromanyl-2-morpholin-4-ylcarbonyl-6-nitro-phenyl)amino]cyclohexyl]-2-oxidanylidene-1~{H}-quinoline-4-carboxamide | | Authors: | Nie, T.Q, Su, H.X, Li, M.J, Xu, Y.C. | | Deposit date: | 2024-01-30 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Discovery of Novel Nonpeptidic and Noncovalent Small Molecule 3CL pro Inhibitors as anti-SARS-CoV-2 Drug Candidate.
J.Med.Chem., 67, 2024
|
|
8JCN
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with Compound 58 | | Descriptor: | 1-[3-(diphenoxyphosphorylamino)phenyl]ethanone, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Zhu, Y, Rao, Z. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-15 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | De novo design of SARS-CoV-2 main protease inhibitors with characteristic binding modes.
Structure, 32, 2024
|
|
8JCK
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with Compound 32 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Zhao, Y, Zhu, Y, Rao, Z. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-15 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | De novo design of SARS-CoV-2 main protease inhibitors with characteristic binding modes.
Structure, 32, 2024
|
|
8JCM
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with Compound 55 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, HYDROSULFURIC ACID, ... | | Authors: | Zhao, Y, Zhu, Y, Rao, Z. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-15 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | De novo design of SARS-CoV-2 main protease inhibitors with characteristic binding modes.
Structure, 32, 2024
|
|
8JCO
 
 | |
8JCL
 
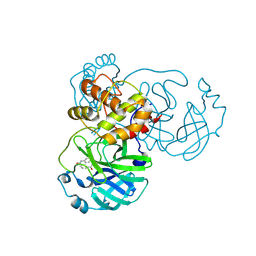 | |
8JCJ
 
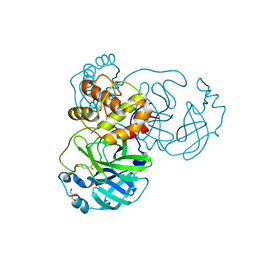 | | The crystal structure of SARS-CoV-2 main protease in complex with Compound 18 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Zhao, Y, Zhu, Y, Rao, Z. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-15 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De novo design of SARS-CoV-2 main protease inhibitors with characteristic binding modes.
Structure, 32, 2024
|
|
6KRE
 
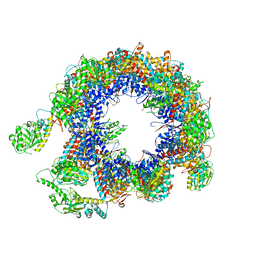 | | TRiC at 0.05 mM ADP-AlFx, Conformation 2, 0.05-C2 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-21 | | Release date: | 2019-09-18 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KRD
 
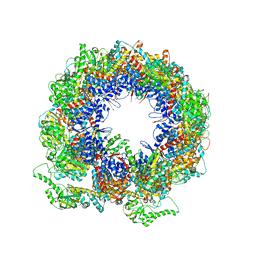 | | TRiC at 0.05 mM ADP-AlFx, Conformation 4, 0.05-C4 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-21 | | Release date: | 2019-09-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.38 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KS8
 
 | | TRiC at 0.1 mM ADP-AlFx, Conformation 4, 0.1-C4 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-23 | | Release date: | 2019-09-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KS7
 
 | | TRiC at 0.1 mM ADP-AlFx, Conformation 1, 0.1-C1 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-23 | | Release date: | 2019-09-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.62 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
