2OYS
 
 | | Crystal Structure of SP1951 protein from Streptococcus pneumoniae in complex with FMN, Northeast Structural Genomics Target SpR27 | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Hypothetical protein SP1951 | | Authors: | Forouhar, F, Lee, I, Vorobiev, S.M, Janjua, H, Satterwhite, R, Liu, J, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional insights from structural genomics.
J.Struct.Funct.Genom., 8, 2007
|
|
2NV4
 
 | | Crystal structure of UPF0066 protein AF0241 in complex with S-adenosylmethionine. Northeast Structural Genomics Consortium target GR27 | | Descriptor: | ACETATE ION, S-ADENOSYLMETHIONINE, UPF0066 protein AF_0241 | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Vorobiev, S.M, Fang, Y, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-10 | | Release date: | 2006-11-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional insights from structural genomics.
J.Struct.Funct.Genom., 8, 2007
|
|
5CIR
 
 | | Crystal structure of death receptor 4 (DR4; TNFFRSF10A) bound to TRAIL (TNFSF10) | | Descriptor: | CHLORIDE ION, Tumor necrosis factor ligand superfamily member 10, Tumor necrosis factor receptor superfamily member 10A, ... | | Authors: | Sheriff, S. | | Deposit date: | 2015-07-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the death receptor 4-TNF-related apoptosis-inducing ligand (DR4-TRAIL) complex.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
2HD3
 
 | | Crystal Structure of the Ethanolamine Utilization Protein EutN from Escherichia coli, NESG Target ER316 | | Descriptor: | Ethanolamine utilization protein eutN | | Authors: | Forouhar, F, Zhang, W, Jayaraman, S, Zhao, L, Jiang, M, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-19 | | Release date: | 2006-08-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
1SQS
 
 | | X-Ray Crystal Structure Protein SP1951 of Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR27. | | Descriptor: | L(+)-TARTARIC ACID, conserved hypothetical protein | | Authors: | Forouhar, F, Lee, I, Vorobiev, S.M, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-19 | | Release date: | 2004-03-30 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM, 8, 2007
|
|
1TM0
 
 | | Crystal Structure of the putative proline racemase from Brucella melitensis, Northeast Structural Genomics Target LR31 | | Descriptor: | PROLINE RACEMASE | | Authors: | Forouhar, F, Chen, Y, Xiao, R, Ho, C.K, Ma, L.-C, Cooper, B, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-10 | | Release date: | 2004-06-29 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
1RTY
 
 | | Crystal Structure of Bacillus subtilis YvqK, a putative ATP-binding Cobalamin Adenosyltransferase, The North East Structural Genomics Target SR128 | | Descriptor: | PHOSPHATE ION, yvqk protein | | Authors: | Forouhar, F, Lee, I, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-10 | | Release date: | 2003-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
7U4P
 
 | |
8BSE
 
 | | CRYSTAL STRUCTURE OF SARS-COV-2 RECEPTOR BINDING DOMAIN (RBD) in complex with 1D1 Fab | | Descriptor: | 1D1 FAB HEAVY CHAIN, 1D1 FAB LIGHT CHAIN, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Welin, M, Kimbung, Y.R, Focht, D, Pisitkun, T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Efficacy of the combination of monoclonal antibodies against the SARS-CoV-2 Beta and Delta variants.
Plos One, 18, 2023
|
|
8BSF
 
 | | CRYSTAL STRUCTURE OF SARS-COV-2 RECEPTOR BINDING DOMAIN (RBD-beta variant) in complex with 3D2 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3D2 FAB HEAVY CHAIN, 3D2 FAB LIGHT CHAIN, ... | | Authors: | Welin, M, Kimbung, Y.R, Focht, D, Pisitkun, T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Efficacy of the combination of monoclonal antibodies against the SARS-CoV-2 Beta and Delta variants.
Plos One, 18, 2023
|
|
7XR0
 
 | | Crystal structure of T2R-TTL-27a complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloranyl-5-fluoranyl-N-(4-methoxyphenyl)-N-methyl-quinazolin-4-amine, CALCIUM ION, ... | | Authors: | Lun, T, Wu, C.Y. | | Deposit date: | 2022-05-09 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of T2R-TTL-27a complex
To Be Published
|
|
7XR1
 
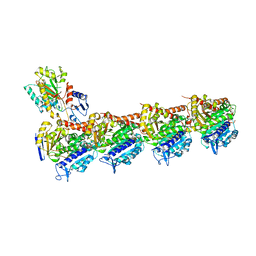 | | Crystal structure of T2R-TTL-3a complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloranyl-6-fluoranyl-N-(4-methoxyphenyl)-N-methyl-quinazolin-4-amine, CALCIUM ION, ... | | Authors: | Lun, T, Wu, C.Y. | | Deposit date: | 2022-05-09 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of T2R-TTL-3a complex
To Be Published
|
|
7CAJ
 
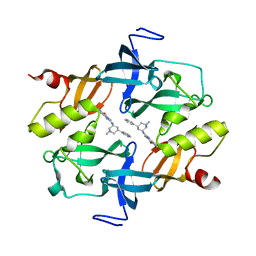 | | Crystal structure of SETDB1 Tudor domain in complexed with Compound 2. | | Descriptor: | 3-methyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]-5H-pyrrolo[3,2-d]pyrimidin-4-one, Histone-lysine N-methyltransferase SETDB1 | | Authors: | Guo, Y.P, Liang, X, Xin, M, Luyi, H, Chengyong, W, Yang, S.Y. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
