5I8I
 
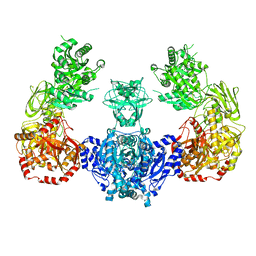 | |
6QVJ
 
 | | HsCKK (human CAMSAP1) decorated 14pf taxol-GDP microtubule | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-03-02 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
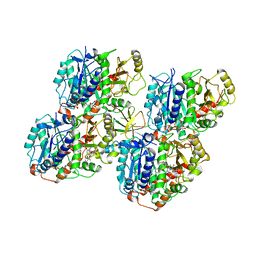
6QUY
 
 | | NgCKK (N.Gruberi CKK) decorated 13pf taxol-GDP microtubule | | Descriptor: | CKK domain protein, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
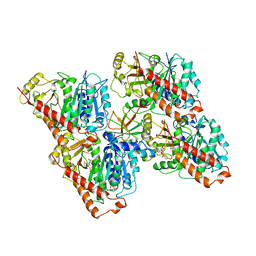
6QVE
 
 | | NgCKK (Naegleria Gruberi CKK) decorated 14pf taxol-GDP microtubule | | Descriptor: | Beta1-tubulin, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-03-01 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
3O2T
 
 | |
4ISS
 
 | |
2A99
 
 | | Crystal structure of recombinant chicken sulfite oxidase at resting state | | Descriptor: | CHLORIDE ION, GLYCEROL, MOLYBDENUM ATOM, ... | | Authors: | Karakas, E, Wilson, H.L, Graf, T.N, Xiang, S, Jaramillo-Busquets, S, Rajagopalan, K.V, Kisker, C. | | Deposit date: | 2005-07-11 | | Release date: | 2005-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural insights into sulfite oxidase deficiency
J.Biol.Chem., 280, 2005
|
|
2A9A
 
 | | Crystal structure of recombinant chicken sulfite oxidase with the bound product, sulfate, at the active site | | Descriptor: | MOLYBDENUM ATOM, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, SULFATE ION, ... | | Authors: | Karakas, E, Wilson, H.L, Graf, T.N, Xiang, S, Jaramillo-Busquets, S, Rajagopalan, K.V, Kisker, C. | | Deposit date: | 2005-07-11 | | Release date: | 2005-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural insights into sulfite oxidase deficiency
J.Biol.Chem., 280, 2005
|
|
2A9C
 
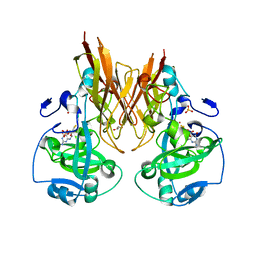 | | Crystal structure of R138Q mutant of recombinant chicken sulfite oxidase with the bound product, sulfate, at the active site | | Descriptor: | GLYCEROL, MOLYBDENUM ATOM, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, ... | | Authors: | Karakas, E, Wilson, H.L, Graf, T.N, Xiang, S, Jaramillo-Busquets, S, Rajagopalan, K.V, Kisker, C. | | Deposit date: | 2005-07-11 | | Release date: | 2005-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Structural insights into sulfite oxidase deficiency
J.Biol.Chem., 280, 2005
|
|
2A9D
 
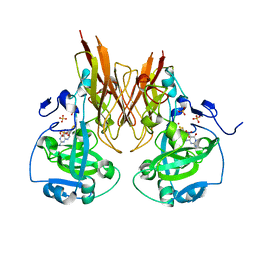 | | Crystal structure of recombinant chicken sulfite oxidase with Arg at residue 161 | | Descriptor: | MOLYBDENUM ATOM, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, SULFATE ION, ... | | Authors: | Karakas, E, Wilson, H.L, Graf, T.N, Xiang, S, Jaramillo-Busquets, S, Rajagopalan, K.V, Kisker, C. | | Deposit date: | 2005-07-11 | | Release date: | 2005-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural insights into sulfite oxidase deficiency
J.Biol.Chem., 280, 2005
|
|
2A9B
 
 | | Crystal structure of R138Q mutant of recombinant sulfite oxidase at resting state | | Descriptor: | CHLORIDE ION, MOLYBDENUM ATOM, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, ... | | Authors: | Karakas, E, Wilson, H.L, Graf, T.N, Xiang, S, Jaramillo-Busquets, S, Rajagopalan, K.V, Kisker, C. | | Deposit date: | 2005-07-11 | | Release date: | 2005-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural insights into sulfite oxidase deficiency
J.Biol.Chem., 280, 2005
|
|
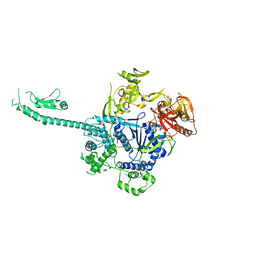
3PIF
 
 | |
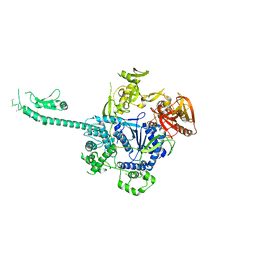
3PIE
 
 | |
6IWW
 
 | | Cryo-EM structure of the S. typhimurium oxaloacetate decarboxylase beta-gamma sub-complex | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Oxaloacetate decarboxylase beta chain, Probable oxaloacetate decarboxylase gamma chain | | Authors: | Xu, X, Shi, H, Zhang, X, Xiang, S. | | Deposit date: | 2018-12-08 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into sodium transport by the oxaloacetate decarboxylase sodium pump.
Elife, 9, 2020
|
|
6IVA
 
 | |
3VA7
 
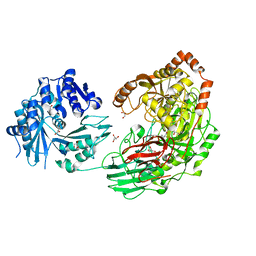 | | Crystal structure of the Kluyveromyces lactis Urea Carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, GLYCEROL, KLLA0E08119p, ... | | Authors: | Fan, C, Xiang, S. | | Deposit date: | 2011-12-29 | | Release date: | 2012-02-01 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of urea carboxylase provides insights into the carboxyltransfer reaction
J.Biol.Chem., 287, 2012
|
|
2MKC
 
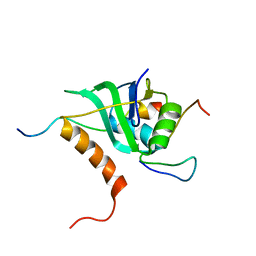 | | Cooperative Structure of the Heterotrimeric pre-mRNA Retention and Splicing Complex | | Descriptor: | Pre-mRNA leakage protein 1, Pre-mRNA-splicing factor CWC26, U2 snRNP component IST3 | | Authors: | Wysoczanski, P, Schneider, C, Xiang, S, Munari, F, Trowitzsch, S, Wahl, M.C, Luhrmann, R, Becker, S, Zweckstetter, M. | | Deposit date: | 2014-02-04 | | Release date: | 2014-09-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cooperative structure of the heterotrimeric pre-mRNA retention and splicing complex.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5D06
 
 | |
5D0F
 
 | |
4IST
 
 | |
7W76
 
 | | Crystal structure of the K. lactis Bre1 RBD in complex with Rad6, crystal form II | | Descriptor: | E3 ubiquitin-protein ligase BRE1, GLYCEROL, SULFATE ION, ... | | Authors: | Shi, M, Zhao, J, Xiang, S. | | Deposit date: | 2021-12-03 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis for the Rad6 activation by the Bre1 N-terminal domain.
Elife, 12, 2023
|
|
7W75
 
 | |
5ZT0
 
 | |
5ZQV
 
 | |
2NRO
 
 | | MoeA K279Q | | Descriptor: | Molybdopterin biosynthesis protein moeA | | Authors: | Nicolas, J, Xiang, S, Schindelin, H, Rajagopalan, K.V. | | Deposit date: | 2006-11-02 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutational Analysis of Escherichia coli MoeA: Two Functional Activities Map to the Active Site Cleft.
Biochemistry, 46, 2007
|
|
