7PJH
 
 | | Crystal structure of the human spliceosomal maturation factor AAR2 bound to the RNAse H domain of PRPF8 | | Descriptor: | Pre-mRNA-processing-splicing factor 8, Protein AAR2 homolog | | Authors: | Preussner, M, Santos, K, Heroven, A.C, Alles, J, Heyd, F, Wahl, M.C, Weber, G. | | Deposit date: | 2021-08-24 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and functional investigation of the human snRNP assembly factor AAR2 in complex with the RNase H-like domain of PRPF8.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7OS2
 
 | | Cryo-EM structure of Brr2 in complex with Jab1/MPN and C9ORF78 | | Descriptor: | Pre-mRNA-processing-splicing factor 8, Telomere length and silencing protein 1 homolog, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Bergfort, A, Hilal, T, Weber, G, Wahl, M.C. | | Deposit date: | 2021-06-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | The intrinsically disordered TSSC4 protein acts as a helicase inhibitor, placeholder and multi-interaction coordinator during snRNP assembly and recycling.
Nucleic Acids Res., 50, 2022
|
|
5LM7
 
 | | Crystal structure of the lambda N-Nus factor complex | | Descriptor: | 30S ribosomal protein S10, Antitermination protein N, N utilization substance protein B homolog, ... | | Authors: | Said, N, Santos, K, Weber, G, Wahl, M.C. | | Deposit date: | 2016-07-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis for lambda N-dependent processive transcription antitermination.
Nat Microbiol, 2, 2017
|
|
4RZ9
 
 | | Pre-mRNA-splicing factor 38A AS 1-179 | | Descriptor: | Pre-mRNA-splicing factor 38A | | Authors: | Schuetze, T, Weber, G, Wahl, M.C. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | The N-terminal domain of the unusual SR protein hPrp38 is an interaction hub in the spliceosome
To be Published
|
|
6QG9
 
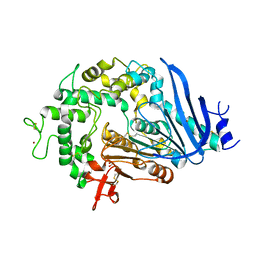 | | Crystal structure of Ideonella sakaiensis MHETase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
6QGC
 
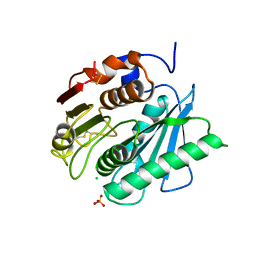 | | PETase from Ideonella sakaiensis without ligand | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
6QGA
 
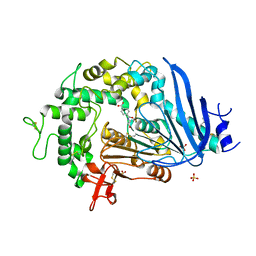 | | Crystal structure of Ideonella sakaiensis MHETase bound to the non-hydrolyzable ligand MHETA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-hydroxyethylcarbamoyl)benzoic acid, CALCIUM ION, ... | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
6QGB
 
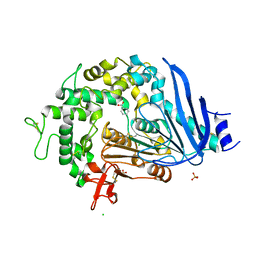 | | Crystal structure of Ideonella sakaiensis MHETase bound to benzoic acid | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BENZOIC ACID, CALCIUM ION, ... | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
7PX3
 
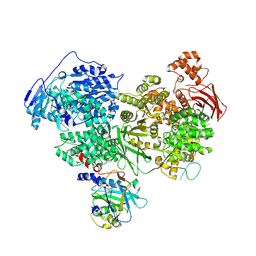 | | Structure of U5 snRNP assembly and recycling factor TSSC4 in complex with BRR2 and Jab1 domain of PRPF8 | | Descriptor: | Pre-mRNA-processing-splicing factor 8, Protein TSSC4, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Bergfort, A, Kuropka, B, Ilik, I.A, Freund, C, Aktas, T, Hilal, T, Weber, G, Wahl, M.C. | | Deposit date: | 2021-10-07 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | The intrinsically disordered TSSC4 protein acts as a helicase inhibitor, placeholder and multi-interaction coordinator during snRNP assembly and recycling.
Nucleic Acids Res., 50, 2022
|
|
5LM9
 
 | | Structure of E. coli NusA | | Descriptor: | MAGNESIUM ION, SULFATE ION, Transcription termination/antitermination protein NusA | | Authors: | Said, N, Weber, G, Santos, K, Wahl, M.C. | | Deposit date: | 2016-07-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | Structural basis for lambda N-dependent processive transcription antitermination.
Nat Microbiol, 2, 2017
|
|
5MPY
 
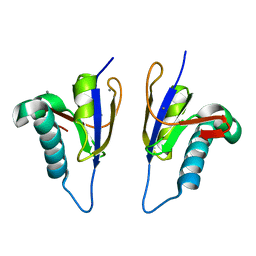 | | Crystal structure of Arabidopsis thaliana RNA editing factor MORF9 | | Descriptor: | CALCIUM ION, Multiple organellar RNA editing factor 9, chloroplastic | | Authors: | Haag, S, Schindler, M, Berndt, L, Brennicke, A, Takenaka, M, Weber, G. | | Deposit date: | 2016-12-19 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Crystal structures of the Arabidopsis thaliana organellar RNA editing factors MORF1 and MORF9.
Nucleic Acids Res., 45, 2017
|
|
5MPX
 
 | | Crystal structure of Arabidopsis thaliana RNA editing factor MORF1, space group P2(1) | | Descriptor: | Multiple organellar RNA editing factor 1, mitochondrial, SULFATE ION | | Authors: | Haag, S, Schindler, M, Berndt, L, Brennicke, A, Takenaka, M, Weber, G. | | Deposit date: | 2016-12-19 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Crystal structures of the Arabidopsis thaliana organellar RNA editing factors MORF1 and MORF9.
Nucleic Acids Res., 45, 2017
|
|
5MPW
 
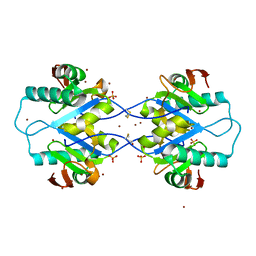 | | Crystal structure of Arabidopsis thaliana RNA editing factor MORF1 | | Descriptor: | BROMIDE ION, Multiple organellar RNA editing factor 1, mitochondrial, ... | | Authors: | Haag, S, Schindler, M, Berndt, L, Brennicke, A, Takenaka, M, Weber, G. | | Deposit date: | 2016-12-19 | | Release date: | 2017-02-22 | | Last modified: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Crystal structures of the Arabidopsis thaliana organellar RNA editing factors MORF1 and MORF9.
Nucleic Acids Res., 45, 2017
|
|
3HD7
 
 | | HELICAL EXTENSION OF THE NEURONAL SNARE COMPLEX INTO THE MEMBRANE, spacegroup C 1 2 1 | | Descriptor: | SULFATE ION, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | Authors: | Stein, A, Weber, G, Wahl, M.C, Jahn, R. | | Deposit date: | 2009-05-07 | | Release date: | 2009-07-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Helical extension of the neuronal SNARE complex into the membrane
Nature, 460, 2009
|
|
3IPD
 
 | | Helical extension of the neuronal SNARE complex into the membrane, spacegroup I 21 21 21 | | Descriptor: | Synaptosomal-associated protein 25, Syntaxin-1A, Vesicle-associated membrane protein 2 | | Authors: | Stein, A, Weber, G, Wahl, M.C, Jahn, R. | | Deposit date: | 2009-08-17 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Helical extension of the neuronal SNARE complex into the membrane
Nature, 460, 2009
|
|
3ST2
 
 | | Dreiklang - equilibrium state | | Descriptor: | Dreiklang, PHOSPHATE ION | | Authors: | Brakemann, T, Weber, G, Andresen, M, Stiel, A.C, Jakobs, S, Wahl, M.C. | | Deposit date: | 2011-07-08 | | Release date: | 2011-09-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A reversibly photoswitchable GFP-like protein with fluorescence excitation decoupled from switching.
Nat.Biotechnol., 29, 2011
|
|
4F92
 
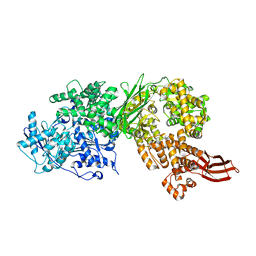 | | Brr2 Helicase Region S1087L | | Descriptor: | SULFANILAMIDE, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Santos, K.F, Jovin, S.M, Weber, G, Pena, V, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.662 Å) | | Cite: | Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ST3
 
 | | Dreiklang - off state | | Descriptor: | Dreiklang, PHOSPHATE ION | | Authors: | Brakemann, T, Weber, G, Andresen, M, Stiel, A.C, Jakobs, S, Wahl, M.C. | | Deposit date: | 2011-07-08 | | Release date: | 2011-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | A reversibly photoswitchable GFP-like protein with fluorescence excitation decoupled from switching.
Nat.Biotechnol., 29, 2011
|
|
3ST4
 
 | | Dreiklang - on state | | Descriptor: | Dreiklang, PHOSPHATE ION | | Authors: | Brakemann, T, Weber, G, Andresen, M, Stiel, A.C, Jakobs, S, Wahl, M.C. | | Deposit date: | 2011-07-08 | | Release date: | 2011-09-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A reversibly photoswitchable GFP-like protein with fluorescence excitation decoupled from switching.
Nat.Biotechnol., 29, 2011
|
|
4F93
 
 | | Brr2 Helicase Region S1087L, Mg-ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Santos, K.F, Jovin, S.M, Weber, G, Pena, V, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F91
 
 | | Brr2 Helicase Region | | Descriptor: | U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Santos, K.F, Jovin, S.M, Weber, G, Pena, V, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ELV
 
 | | Crystal Structure of Full-Length Yeast Pml1p | | Descriptor: | Pre-mRNA leakage protein 1, SULFATE ION | | Authors: | Trowitzsch, S, Weber, G, L hrmann, R, Wahl, M.C. | | Deposit date: | 2008-09-23 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the Pml1p subunit of the yeast precursor mRNA retention and splicing complex.
J.Mol.Biol., 385, 2009
|
|
3ELS
 
 | | Crystal Structure of Yeast Pml1p, Residues 51-204 | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Trowitzsch, S, Weber, G, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2008-09-23 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Pml1p subunit of the yeast precursor mRNA retention and splicing complex.
J.Mol.Biol., 385, 2009
|
|
2IOV
 
 | | Bright-state structure of the reversibly switchable fluorescent protein Dronpa | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Stiel, A.C, Trowitzsch, S, Weber, G, Andresen, M, Eggeling, C, Hell, S.W, Jakobs, S, Wahl, M.C. | | Deposit date: | 2006-10-11 | | Release date: | 2006-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 A bright-state structure of the reversibly switchable fluorescent protein Dronpa guides the generation of fast switching variants
Biochem.J., 402, 2007
|
|
7E30
 
 | | Crystal structure of a novel alpha/beta hydrolase in apo form in complex with citrate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CITRIC ACID, SULFATE ION, ... | | Authors: | Gao, J, Han, X, Zheng, Y.Y, Liu, W.D. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
