6UCO
 
 | |
6UCP
 
 | |
6E5H
 
 | |
6WYA
 
 | | RTX (Reverse Transcription Xenopolymerase) in complex with a DNA duplex and dAMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Choi, W.S, He, P, Pothukuchy, A, Gollihar, J, Ellington, A.D, Yang, W. | | Deposit date: | 2020-05-12 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | How a B family DNA polymerase has been evolved to copy RNA.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WYB
 
 | | RTX (Reverse Transcription Xenopolymerase) in complex with an RNA/DNA hybrid | | Descriptor: | 1,2-ETHANEDIOL, 3,3',3''-phosphanetriyltripropanoic acid, DNA polymerase, ... | | Authors: | Choi, W.S, He, P, Pothukuchy, A, Gollihar, J, Ellington, A.D, Yang, W. | | Deposit date: | 2020-05-12 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | How a B family DNA polymerase has been evolved to copy RNA.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6E5I
 
 | |
6H2B
 
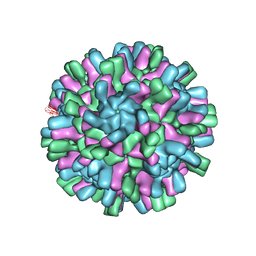 | | Structure of the Macrobrachium rosenbergii Nodavirus | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Ho, K.H, Gabrielsen, M, Beh, P.L, Kueh, C.L, Thong, Q.X, Streetley, J, Tan, W.S, Bhella, D. | | Deposit date: | 2018-07-13 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the Macrobrachium rosenbergii nodavirus: A new genus within the Nodaviridae?
PLoS Biol., 16, 2018
|
|
7E8Z
 
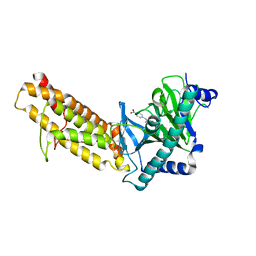 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with SS81 | | Descriptor: | 2-[[6-[(4-nitrophenyl)amino]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, ZINC ION | | Authors: | Tam, N.Y, Ng, Y.M, Shishodia, S, McDonough, M.A, Schofield, C.J, Aik, W.S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-Based Design of Selective Fat Mass and Obesity Associated Protein (FTO) Inhibitors.
J.Med.Chem., 64, 2021
|
|
7FHC
 
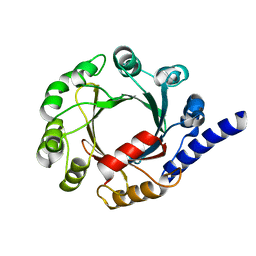 | | Structure of prenyltransferase mutant V49W from Streptomyces sp. (strain CL190) | | Descriptor: | Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7FHB
 
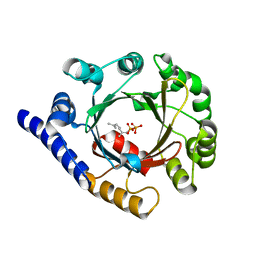 | | Structure of prenyltransferase from Streptomyces sp. (strain CL190) with bound GPP | | Descriptor: | GERANYL DIPHOSPHATE, Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7FHD
 
 | | Structure of prenyltransferase mutant Y288P from Streptomyces sp. (strain CL190) | | Descriptor: | Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7FHE
 
 | | Structure of prenyltransferase mutant Q295F from Streptomyces sp. (strain CL190) | | Descriptor: | Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7FHF
 
 | | Structure of prenyltransferase mutant V49W/Y288F/Q295F from Streptomyces sp. (strain CL190) | | Descriptor: | Prenyltransferase | | Authors: | Xue, B, Lim, K.J.H, Hartono, Y.D, Go, M.D.K, Fan, H, Yew, W.S. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Engineering of Prenyltransferase NphB for High-Yield and Regioselective Cannabinoid Production.
Acs Catalysis, 12, 2022
|
|
7DWM
 
 | | Crystal structure of the phage VqmA-DPO complex | | Descriptor: | 3,5-dimethylpyrazin-2-ol, Transcriptional regulator | | Authors: | Gu, Y, Yang, W.S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Understanding the mechanism of asymmetric gene regulation determined by the VqmA of vibriophage.
Biochem.Biophys.Res.Commun., 558, 2021
|
|
8JZD
 
 | | Crystal structure of Escherichia coli NarJ in complex with the signal peptide of E. coli NarG | | Descriptor: | Nitrate reductase molybdenum cofactor assembly chaperone NarJ, Respiratory nitrate reductase 1 alpha chain | | Authors: | Song, W.S, Kim, J.H, Namgung, B, Cho, H.Y, Oh, H.B, Yoon, S.I. | | Deposit date: | 2023-07-05 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Complementary hydrophobic interaction of the redox enzyme maturation protein NarJ with the signal peptide of the respiratory nitrate reductase NarG.
Int.J.Biol.Macromol., 262, 2024
|
|
8JZC
 
 | | Crystal structure of Geobacillus stearothermophilus NarJ | | Descriptor: | Nitrate reductase molybdenum cofactor assembly chaperone | | Authors: | Song, W.S, Kim, J.H, Namgung, B, Cho, H.Y, Oh, H.B, Yoon, S.I. | | Deposit date: | 2023-07-05 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Complementary hydrophobic interaction of the redox enzyme maturation protein NarJ with the signal peptide of the respiratory nitrate reductase NarG.
Int.J.Biol.Macromol., 262, 2024
|
|
1HKG
 
 | |
5GY2
 
 | |
5H5V
 
 | |
4A1U
 
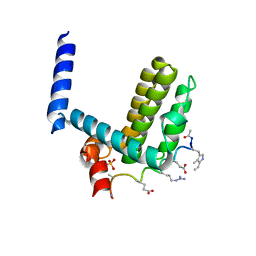 | | Crystal structure of alpha-beta-foldamer 2c in complex with Bcl-xL | | Descriptor: | ALPHA-BETA-FOLDAMER 2C, BCL-2-LIKE PROTEIN 1, CHLORIDE ION, ... | | Authors: | Boersma, M.D, Haase, H.S, Kaufman, K.J, Horne, W.S, Lee, E.F, Clarke, O.B, Smith, B.J, Colman, P.M, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2011-09-20 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Evaluation of Diverse Alpha/Beta-Backbone Patterns for Functional Alpha-Helix Mimicry: Analogues of the Bim Bh3 Domain.
J.Am.Chem.Soc., 134, 2012
|
|
4A1W
 
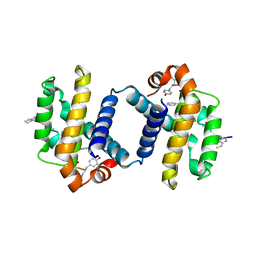 | | Crystal structure of alpha-beta foldamer 4c in complex with Bcl-xL | | Descriptor: | ALPHA-BETA-FOLDAMER 2C, BCL-2-LIKE PROTEIN 1 | | Authors: | Boersma, M.D, Haase, H.S, Kaufman, K.J, Horne, W.S, Lee, E.F, Clarke, O.B, Smith, B.J, Colman, P.M, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2011-09-20 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Evaluation of Diverse Alpha/Beta-Backbone Patterns for Functional Alpha-Helix Mimicry: Analogues of the Bim Bh3 Domain.
J.Am.Chem.Soc., 134, 2012
|
|
5HFY
 
 | | Backbone Modifications in the Protein GB1 Helix: beta-2-Ala24, beta-3-Lys28, beta-3-Lys31, beta-3-Asn35 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Tavenor, N.A, Reinert, Z.E, Lengyel, G.A, Griffith, B.D, Horne, W.S. | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Comparison of design strategies for alpha-helix backbone modification in a protein tertiary fold.
Chem.Commun.(Camb.), 52, 2016
|
|
5HG2
 
 | | Backbone Modifications in the Protein GB1 Helix: beta-3-Ala24, beta-3-Lys28, beta-3-Lys31, beta-2-Asn35 | | Descriptor: | GLYCEROL, Immunoglobulin G-binding protein G, MAGNESIUM ION | | Authors: | Tavenor, N.A, Reinert, Z.E, Lengyel, G.A, Griffith, B.D, Horne, W.S. | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of design strategies for alpha-helix backbone modification in a protein tertiary fold.
Chem.Commun.(Camb.), 52, 2016
|
|
5H5W
 
 | |
5H5T
 
 | |
