5O5X
 
 | | Crystal structure of Thermococcus litoralis ADP-dependent glucokinase (GK) | | Descriptor: | ADP-dependent glucokinase,ADP-dependent glucokinase,ADP-dependent glucokinase, GLYCEROL, SULFATE ION | | Authors: | Herrera-Morande, A, Castro-Fernandez, V, Merino, F, Ramirez-Sarmiento, C.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2017-06-02 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Protein topology determines substrate-binding mechanism in homologous enzymes.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
6RJN
 
 | | Crystal structure of a Fungal Catalase at 2.3 Angstroms | | Descriptor: | CHLORIDE ION, Catalase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gomez, S, Navas-Yuste, S, Payne, A.M, Rivera, W, Lopez-Estepa, M, Brangbour, C, Fulla, D, Juanhuix, J, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2019-04-28 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Peroxisomal catalases from the yeasts Pichia pastoris and Kluyveromyces lactis as models for oxidative damage in higher eukaryotes.
Free Radic. Biol. Med., 141, 2019
|
|
6TJP
 
 | | Crystal structure of T7 bacteriophage portal protein, 13mer, closed valve - P212121 | | Descriptor: | Portal protein | | Authors: | Fabrega-Ferrer, M, Cuervo, A, Fernandez, F.J, Machon, C, Perez-Luque, R, Pous, J, Vega, M.C, Carrascosa, J.L, Coll, M. | | Deposit date: | 2019-11-26 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | Using a partial atomic model from medium-resolution cryo-EM to solve a large crystal structure.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6QXM
 
 | | Cryo-EM structure of T7 bacteriophage portal protein, 12mer, open valve | | Descriptor: | Portal protein | | Authors: | Fabrega-Ferrer, M, Cuervo, A, Machon, C, Fernandez, F.J, Perez-Luque, R, Pous, J, Vega, M.C, Carrascosa, J.L, Coll, M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism.
Nat Commun, 10, 2019
|
|
6QWP
 
 | | Crystal structure of T7 bacteriophage portal protein, 13mer, closed valve | | Descriptor: | Portal protein | | Authors: | Fabrega-Ferrer, M, Cuervo, A, Machon, C, Fernandez, F.J, Perez-Luque, R, Pous, J, Vega, M.C, Carrascosa, J.L, Coll, M. | | Deposit date: | 2019-03-06 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism.
Nat Commun, 10, 2019
|
|
5LD5
 
 | | Crystal structure of a bacterial dehydrogenase at 2.19 Angstroms resolution | | Descriptor: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Querol-Garcia, J, Fernandez, F.J, Gomez, S, Fulla, D, Juanhuix, J, Vega, M.C. | | Deposit date: | 2016-06-23 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1906 Å) | | Cite: | Crystal Structure of Glyceraldehyde-3-Phosphate Dehydrogenase from the Gram-Positive Bacterial Pathogen A. vaginae, an Immunoevasive Factor that Interacts with the Human C5a Anaphylatoxin.
Front Microbiol, 8, 2017
|
|
6QX5
 
 | | Crystal structure of T7 bacteriophage portal protein, 12mer, closed valve | | Descriptor: | Portal protein | | Authors: | Fabrega-Ferrer, M, Cuervo, A, Machon, C, Fernandez, F.J, Perez-Luque, R, Pous, J, Vega, M.C, Carrascosa, J.L, Coll, M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism.
Nat Commun, 10, 2019
|
|
6RJR
 
 | | Crystal structure of a Fungal Catalase at 1.9 Angstrom | | Descriptor: | CHLORIDE ION, Catalase, GLYCEROL, ... | | Authors: | Gomez, S, Navas-Yuste, S, Payne, A.M, Rivera, W, Lopez-Estepa, M, Brangbour, C, Fulla, D, Juanhuix, J, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2019-04-29 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Peroxisomal catalases from the yeasts Pichia pastoris and Kluyveromyces lactis as models for oxidative damage in higher eukaryotes.
Free Radic. Biol. Med., 141, 2019
|
|
6TAI
 
 | | Crystal structure of Escherichia coli Orotate Phosphoribosyltransferase with an empty active site at 1.55 Angstrom resolution | | Descriptor: | ACETATE ION, GLYCEROL, Orotate phosphoribosyltransferase | | Authors: | Navas-Yuste, S, Lopez-Estepa, M, Gomez, S, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Elucidating the Catalytic Reaction Mechanism of Orotate Phosphoribosyltransferase by Means of X-ray Crystallography and Computational Simulations
Acs Catalysis, 10, 2020
|
|
6TAJ
 
 | | Crystal structure of Escherichia coli Orotate Phosphoribosyltransferase in complex with Orotic acid 1.60 Angstrom resolution | | Descriptor: | GLYCEROL, OROTIC ACID, Orotate phosphoribosyltransferase | | Authors: | Navas-Yuste, S, Lopez-Estepa, M, Gomez, S, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Elucidating the Catalytic Reaction Mechanism of Orotate Phosphoribosyltransferase by Means of X-ray Crystallography and Computational Simulations
Acs Catalysis, 10, 2020
|
|
6TAK
 
 | | Crystal structure of Escherichia coli Orotate Phosphoribosyltransferase in complex with Orotic acid and Sulfate at 1.25 Angstrom resolution | | Descriptor: | GLYCEROL, OROTIC ACID, Orotate phosphoribosyltransferase, ... | | Authors: | Navas-Yuste, S, Lopez-Estepa, M, Gomez, S, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Elucidating the Catalytic Reaction Mechanism of Orotate Phosphoribosyltransferase by Means of X-ray Crystallography and Computational Simulations
Acs Catalysis, 10, 2020
|
|
2XQ1
 
 | | Crystal structure of peroxisomal catalase from the yeast Hansenula polymorpha | | Descriptor: | PEROXISOMAL CATALASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Penya-Soler, E, Vega, M.C, Wilmanns, M, Williams, C.P. | | Deposit date: | 2010-08-31 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Features of Peroxisomal Catalase from the Yeast Hansenula Polymorpha
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2WYL
 
 | | Apo structure of a metallo-b-lactamase | | Descriptor: | FORMYL GROUP, GLYCEROL, L-ASCORBATE-6-PHOSPHATE LACTONASE ULAG | | Authors: | Garces, F, Fernandez, F.J, Penya-Soler, E, Aguilar, J, Baldoma, L, Coll, M, Badia, J, Vega, M.C. | | Deposit date: | 2009-11-16 | | Release date: | 2010-04-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Molecular Architecture of the Mn(2+)Dependent Lactonase Ulag Reveals an Rnase-Like Metallo-Beta-Lactamase Fold and a Novel Quaternary Structure.
J.Mol.Biol., 398, 2010
|
|
2WYM
 
 | | Structure of a metallo-b-lactamase | | Descriptor: | CITRATE ANION, GLYCEROL, L-ASCORBATE-6-PHOSPHATE LACTONASE ULAG, ... | | Authors: | Garces, F, Fernandez, F.J, Penya-Soler, E, Aguilar, J, Baldoma, L, Coll, M, Badia, J, Vega, M.C. | | Deposit date: | 2009-11-16 | | Release date: | 2010-04-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Architecture of the Mn(2+)Dependent Lactonase Ulag Reveals an Rnase-Like Metallo-Beta-Lactamase Fold and a Novel Quaternary Structure.
J.Mol.Biol., 398, 2010
|
|
2CDT
 
 | | alpha-SPECTRIN SH3 DOMAIN A56S MUTANT | | Descriptor: | SPECTRIN ALPHA CHAIN | | Authors: | Casares, S, Camara-Artigas, A, Vega, M.C, Lopez-Mayorga, O, Conejero-Lara, F. | | Deposit date: | 2006-01-27 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Cooperative Propagation of Local Stability Changes from Low-Stability and High-Stability Regions in a SH3 Domain.
Proteins: Struct., Funct., Bioinf., 67, 2007
|
|
1JVW
 
 | | TRYPANOSOMA CRUZI MACROPHAGE INFECTIVITY POTENTIATOR (TCMIP) | | Descriptor: | MACROPHAGE INFECTIVITY POTENTIATOR | | Authors: | Pereira, P.J.B, Vega, M.C, Gonzalez-Rey, E, Fernandez-Carazo, R, Macedo-Ribeiro, S, Gomis-Rueth, F.X, Gonzalez, A, Coll, M. | | Deposit date: | 2001-08-31 | | Release date: | 2002-06-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Trypanosoma cruzi macrophage infectivity potentiator has a rotamase core and a highly exposed alpha-helix.
EMBO Rep., 3, 2002
|
|
7AKK
 
 | | Structure of a complement factor-receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3 beta chain, Complement C3b alpha' chain, ... | | Authors: | Fernandez, F.J, Santos-Lopez, J, Martinez-Barricarte, R, Querol-Garcia, J, Navas-Yuste, S, Savko, M, Shepard, W.E, Rodriguez de Cordoba, S, Vega, M.C. | | Deposit date: | 2020-10-01 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.395 Å) | | Cite: | The crystal structure of iC3b-CR3 alpha I reveals a modular recognition of the main opsonin iC3b by the CR3 integrin receptor
Nat Commun, 13, 2022
|
|
8OHA
 
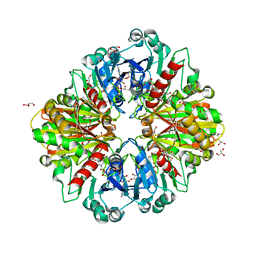 | | Crystal structure of Leptospira interrogans GAPDH | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Navas-Yuste, S, de la Paz, K, Querol-Garcia, J, Gomez-Quevedo, S, Rodriguez de Cordoba, S, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The structure of Leptospira interrogans GAPDH sheds light into an immunoevasion factor that can target the anaphylatoxin C5a of innate immunity.
Front Immunol, 14, 2023
|
|
1N5Z
 
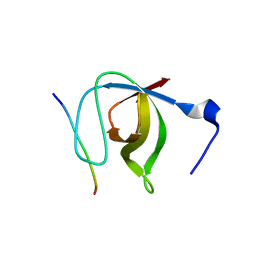 | | Complex structure of Pex13p SH3 domain with a peptide of Pex14p | | Descriptor: | 14-mer peptide from Peroxisomal membrane protein PEX14, Peroxisomal membrane protein PAS20 | | Authors: | Douangamath, A, Filipp, F.V, Klein, A.T.J, Barnett, P, Zou, P, Voorn-Brouwer, T, Vega, M.C, Mayans, O.M, Sattler, M, Distel, B, Wilmanns, M. | | Deposit date: | 2002-11-08 | | Release date: | 2002-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Topography for Independent Binding of alpha-Helical and PPII-Helical Ligands to a Peroxisomal SH3 Domain
MOL.CELL, 10, 2002
|
|
1NQ6
 
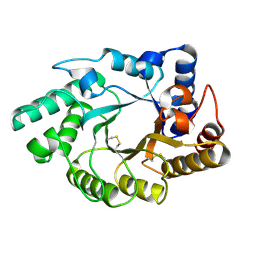 | | Crystal Structure of the catalytic domain of xylanase A from Streptomyces halstedii JM8 | | Descriptor: | MAGNESIUM ION, Xys1 | | Authors: | Canals, A, Vega, M.C, Gomis-Ruth, F.X, Santamaria, R.I, Coll, M. | | Deposit date: | 2003-01-21 | | Release date: | 2004-01-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of xylanase Xys1delta from Streptomyces halstedii.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5FT8
 
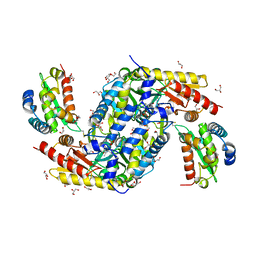 | | Crystal structure of the complex between the cysteine desulfurase CsdA and the sulfur-acceptor CsdE in the persulfurated state at 2.50 Angstroem resolution | | Descriptor: | Cysteine desulfurase CsdA, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Fernandez, F.J, Arda, A, Lopez-Estepa, M, Aranda, J, Penya-Soler, E, Garces, F, Round, A, Campos-Oliva, R, Bruix, M, Coll, M, Tunon, I, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Sulfur Transfer Across Protein-Protein Interfaces: The Cysteine Desulfurase Model System
Acs Catalysis, 6, 2016
|
|
5FT5
 
 | | Crystal structure of the cysteine desulfurase CsdA (persulfurated) from Escherichia coli at 2.384 Angstroem resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Fernandez, F.J, Arda, A, Lopez-Estepa, M, Aranda, J, Penya-Soler, E, Garces, F, Quintana, J.F, Round, A, Campos-Oliva, R, Bruix, M, Coll, M, Tunon, I, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.384 Å) | | Cite: | Mechanism of Sulfur Transfer Across Protein-Protein Interfaces: The Cysteine Desulfurase Model System
Acs Catalysis, 6, 2016
|
|
5FT4
 
 | | Crystal structure of the cysteine desulfurase CsdA from Escherichia coli at 1.996 Angstroem resolution | | Descriptor: | CITRIC ACID, CYSTEINE DESULFURASE CSDA, GLYCEROL, ... | | Authors: | Fernandez, F.J, Arda, A, Lopez-Estepa, M, Aranda, J, Penya-Soler, E, Garces, F, Quintana, J.F, Round, A, Campos-Oliva, R, Bruix, M, Coll, M, Tunon, I, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-12-21 | | Last modified: | 2019-01-02 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | The Mechanism of Sulfur Transfer Across Protein- Protein Interfaces: The Csd Model
Acs Catalysis, 6, 2016
|
|
5FT6
 
 | | Crystal structure of the cysteine desulfurase CsdA (S-sulfonic acid) from Escherichia coli at 2.050 Angstroem resolution | | Descriptor: | CYSTEINE DESULFURASE CSDA, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Fernandez, F.J, Arda, A, Lopez-Estepa, M, Aranda, J, Penya-Soler, E, Garces, F, Round, A, Campos-Oliva, R, Bruix, M, Coll, M, Tunon, I, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Mechanism of Sulfur Transfer Across Protein-Protein Interfaces: The Cysteine Desulfurase Model System
Acs Catalysis, 6, 2016
|
|
4B8R
 
 | | Crystal Structure of Thermococcus litoralis ADP-dependent Glucokinase (GK) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADP-DEPENDENT GLUCOKINASE, ... | | Authors: | Herrera-Morande, A, Rivas-Pardo, J.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2012-08-30 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure, Saxs and Kinetic Mechanism of Hyperthermophilic Adp-Dependent Glucokinase from Thermococcus Litoralis Reveal a Conserved Mechanism for Catalysis.
Plos One, 8, 2013
|
|
