2NNF
 
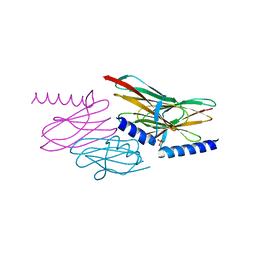 | | Structure of the sulfur carrier protein SoxY from Chlorobium limicola f thiosulfatophilum | | Descriptor: | PHOSPHATE ION, Sulfur covalently-binding protein | | Authors: | Stout, J, Van Driessche, G, Savvides, S.N, Van Beeumen, J. | | Deposit date: | 2006-10-24 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | X-ray crystallographic analysis of the sulfur carrier protein SoxY from Chlorobium limicola f. thiosulfatophilum reveals a tetrameric structure.
Protein Sci., 16, 2007
|
|
1I2S
 
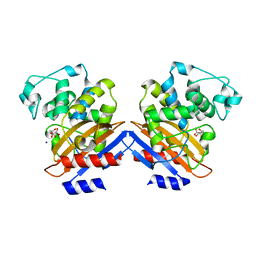 | | BETA-LACTAMASE FROM BACILLUS LICHENIFORMIS BS3 | | Descriptor: | BETA-LACTAMASE, CITRIC ACID, SODIUM ION | | Authors: | Fonze, E, Vanhove, M, Dive, G, Sauvage, E, Frere, J.M, Charlier, P. | | Deposit date: | 2001-02-12 | | Release date: | 2002-03-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the Bacillus licheniformis BS3 class A beta-lactamase and of the
acyl-enzyme adduct formed with cefoxitin
Biochemistry, 41, 2002
|
|
1I2W
 
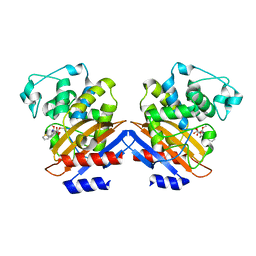 | | BETA-LACTAMASE FROM BACILLUS LICHENIFORMIS BS3 COMPLEXED WITH CEFOXITIN | | Descriptor: | (2R)-5-[(carbamoyloxy)methyl]-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, BETA-LACTAMASE, CARBAMIC ACID | | Authors: | Fonze, E, Vanhove, M, Dive, G, Sauvage, E, Frere, J.M, Charlier, P. | | Deposit date: | 2001-02-12 | | Release date: | 2002-03-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the Bacillus licheniformis BS3 class A beta-lactamase and of the acyl-enzyme adduct formed with cefoxitin
Biochemistry, 41, 2002
|
|
1HIX
 
 | | CRYSTALLOGRAPHIC ANALYSES OF FAMILY 11 ENDO-BETA-1,4-XYLANASE XYL1 FROM STREPTOMYCES SP. S38 | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Wouters, J, Georis, J, Dusart, J, Frere, J.M, Depiereux, E, Charlier, P. | | Deposit date: | 2001-01-05 | | Release date: | 2001-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Analysis of Family 11 Endo-[Beta]-1,4-Xylanase Xyl1 from Streptomyces Sp. S38
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1W79
 
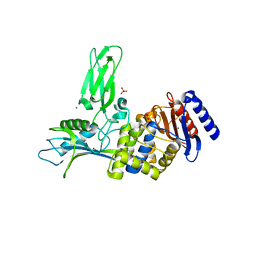 | | Crystal structure of the DD-transpeptidase-carboxypeptidase from Actinomadura R39 | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, MAGNESIUM ION, SULFATE ION | | Authors: | Sauvage, E, Herman, R, Petrella, S, Duez, C, Frere, J.M, Charlier, P. | | Deposit date: | 2004-08-31 | | Release date: | 2005-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Actinomadura R39 DD-peptidase reveals new domains in penicillin-binding proteins.
J. Biol. Chem., 280, 2005
|
|
1NDT
 
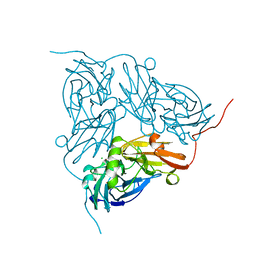 | | NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PROTEIN (NITRITE REDUCTASE) | | Authors: | Dodd, F.E, Vanbeeumen, J, Eady, R.R, Hasnain, S.S. | | Deposit date: | 1998-10-28 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of a blue-copper nitrite reductase in two crystal forms. The nature of the copper sites, mode of substrate binding and recognition by redox partner.
J.Mol.Biol., 282, 1998
|
|
2BE9
 
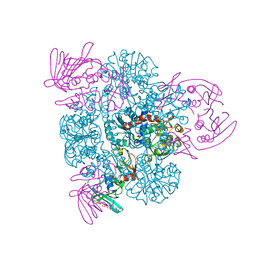 | | Crystal structure of the CTP-liganded (T-State) aspartate transcarbamoylase from the extremely thermophilic archaeon Sulfolobus acidocaldarius | | Descriptor: | Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | De Vos, D, Savvides, S.N, Van Beeumen, J.J. | | Deposit date: | 2005-10-23 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Sulfolobus acidocaldarius aspartate carbamoyltransferase in complex with its allosteric activator CTP.
Biochem.Biophys.Res.Commun., 372, 2008
|
|
6QCL
 
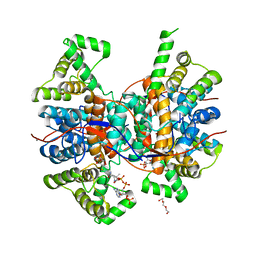 | |
6QFB
 
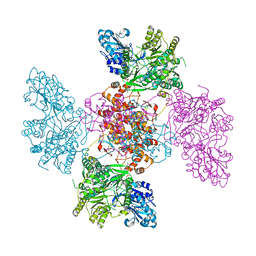 | |
1JMX
 
 | |
1LS9
 
 | |
1JMZ
 
 | | crystal structure of a quinohemoprotein amine dehydrogenase from pseudomonas putida with inhibitor | | Descriptor: | Amine Dehydrogenase, HEME C, NICKEL (II) ION, ... | | Authors: | Satoh, A, Miyahara, I, Hirotsu, K. | | Deposit date: | 2001-07-20 | | Release date: | 2002-01-16 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of quinohemoprotein amine dehydrogenase from Pseudomonas putida. Identification of a novel quinone cofactor encaged by multiple thioether cross-bridges.
J.Biol.Chem., 277, 2002
|
|
8C7L
 
 | |
2B4F
 
 | | Structure Of A Cold-Adapted Family 8 Xylanase in complex with substrate | | Descriptor: | beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, endo-1,4-beta-xylanase | | Authors: | De Vos, D, Collins, T, Savvides, S.N, Feller, G, Van Beeumen, J.J. | | Deposit date: | 2005-09-23 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Oligosaccharide binding in family 8 glycosidases: crystal structures of active-site mutants of the beta-1,4-xylanase pXyl from Pseudoaltermonas haloplanktis TAH3a in complex with substrate and product.
Biochemistry, 45, 2006
|
|
4B5N
 
 | | Crystal structure of oxidized Shewanella Yellow Enzyme 4 (SYE4) | | Descriptor: | FLAVIN MONONUCLEOTIDE, OXIDOREDUCTASE, FMN-BINDING, ... | | Authors: | Elegheert, J, Brige, A, Savvides, S.N. | | Deposit date: | 2012-08-07 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural dissection of Shewanella oneidensis old yellow enzyme 4 bound to a Meisenheimer complex and (nitro)phenolic ligands.
FEBS Lett., 591, 2017
|
|
5K0R
 
 | | Crystal structure of reduced Shewanella Yellow Enzyme 4 (SYE4) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, NAD(P)H:flavin oxidoreductase Sye4, Octadecane | | Authors: | Elegheert, J, Brige, A, Savvides, S.N. | | Deposit date: | 2016-05-17 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural dissection of Shewanella oneidensis old yellow enzyme 4 bound to a Meisenheimer complex and (nitro)phenolic ligands.
FEBS Lett., 591, 2017
|
|
5K1U
 
 | |
1XWQ
 
 | | Structure Of A Cold-Adapted Family 8 Xylanase | | Descriptor: | beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, endo-1,4-beta-xylanase | | Authors: | De Vos, D, Collins, T, Hoyoux, A, Savvides, S.N, Gerday, C, Van Beeumen, J.J, Feller, G. | | Deposit date: | 2004-11-02 | | Release date: | 2005-10-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Study of the active site residues of a glycoside hydrolase family 8 xylanase
J.Mol.Biol., 354, 2005
|
|
5K1K
 
 | | Crystal structure of oxidized Shewanella Yellow Enzyme 4 (SYE4) in complex with p-hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, NAD(P)H:flavin oxidoreductase Sye4, Octadecane, ... | | Authors: | Elegheert, J, Brige, A, Savvides, S.-N. | | Deposit date: | 2016-05-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | Structural dissection of Shewanella oneidensis old yellow enzyme 4 bound to a Meisenheimer complex and (nitro)phenolic ligands.
FEBS Lett., 591, 2017
|
|
5K1M
 
 | |
5K1W
 
 | | Crystal structure of oxidized Shewanella Yellow Enzyme 4 (SYE4) in complex with trinitrophenol | | Descriptor: | FLAVIN MONONUCLEOTIDE, NAD(P)H:flavin oxidoreductase Sye4, Octadecane, ... | | Authors: | Elegheert, J, Brige, A, Savvides, S.N. | | Deposit date: | 2016-05-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural dissection of Shewanella oneidensis old yellow enzyme 4 bound to a Meisenheimer complex and (nitro)phenolic ligands.
FEBS Lett., 591, 2017
|
|
5K1Q
 
 | |
1XWT
 
 | | Structure Of A Cold-Adapted Family 8 Xylanase | | Descriptor: | endo-1,4-beta-xylanase | | Authors: | De Vos, D, Collins, T, Hoyoux, A, Savvides, S.N, Gerday, C, Van Beeumen, J.J, Feller, G. | | Deposit date: | 2004-11-02 | | Release date: | 2005-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Study of the active site residues of a glycoside hydrolase family 8 xylanase
J.Mol.Biol., 354, 2005
|
|
6HXL
 
 | |
6HXO
 
 | |
