2UZU
 
 | | PKA structures of indazole-pyridine series of AKT inhibitors | | Descriptor: | (2S)-1-(1H-INDOL-3-YL)-3-{[5-(3-METHYL-1H-INDAZOL-5-YL)PYRIDIN-3-YL]OXY}PROPAN-2-AMINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Zhu, G.D, Gandhi, V.B, Gong, J, Thomas, S, Woods, K.W, Song, X, Li, T, Diebold, R.B, Luo, Y, Liu, X, Guan, R, Klinghofer, V, Johnson, E.F, Bouska, J, Olson, A, Marsh, K.C, Stoll, V.S, Mamo, M, Polakowski, J, Campbell, T.J, Penning, T.D, Li, Q, Rosenberg, S.H, Giranda, V.L. | | Deposit date: | 2007-05-01 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Syntheses of Potent, Selective, and Orally Bioavailable Indazole-Pyridine Series of Protein Kinase B/Akt Inhibitors with Reduced Hypotension.
J.Med.Chem., 50, 2007
|
|
2UZV
 
 | | PKA structures of indazole-pyridine series of AKT inhibitors | | Descriptor: | (2S)-1-[3-(CYCLOHEXYLMETHOXY)PHENYL]-3-{[5-(3-METHYL-1H-INDAZOL-5-YL)PYRIDIN-3-YL]OXY}PROPAN-2-AMINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA,, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Zhu, G.D, Gandhi, V.B, Gong, J, Thomas, S, Woods, K.W, Song, X, Li, T, Diebold, R.B, Luo, Y, Liu, X, Guan, R, Klinghofer, V, Johnson, E.F, Bouska, J, Olson, A, Marsh, K.C, Stoll, V.S, Mamo, M, Polakowski, J, Campbell, T.J, Penning, T.D, Li, Q, Rosenberg, S.H, Giranda, V.L. | | Deposit date: | 2007-05-01 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Syntheses of Potent, Selective, and Orally Bioavailable Indazole-Pyridine Series of Protein Kinase B/Akt Inhibitors with Reduced Hypotension.
J.Med.Chem., 50, 2007
|
|
2UZW
 
 | | PKA structures of indazole-pyridine series of AKT inhibitors | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.] PYRAZOLE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Zhu, G.D, Gandhi, V.B, Gong, J, Thomas, S, Woods, K.W, Song, X, Li, T, Diebold, R.B, Luo, Y, Liu, X, Guan, R, Klinghofer, V, Johnson, E.F, Bouska, J, Olson, A, Marsh, K.C, Stoll, V.S, Mamo, M, Polakowski, J, Campbell, T.J, Penning, T.D, Li, Q, Rosenberg, S.H, Giranda, V.L. | | Deposit date: | 2007-05-01 | | Release date: | 2007-06-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Syntheses of Potent, Selective, and Orally Bioavailable Indazole-Pyridine Series of Protein Kinase B/Akt Inhibitors with Reduced Hypotension.
J.Med.Chem., 50, 2007
|
|
5TC5
 
 | | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with butylthio-DADMe-Immucillin-A and chloride | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, CHLORIDE ION, S-methyl-5'-thioadenosine phosphorylase | | Authors: | Cameron, S.A, Firestone, R.S, Schramm, V.L, Almo, S.C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | TBA
To be published
|
|
1AQD
 
 | |
5TC8
 
 | | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with methylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cameron, S.A, Firestone, R.S, Schramm, V.L, Almo, S.C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | TBA
To be published
|
|
5TC6
 
 | | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with propylthio-immucillin-A | | Descriptor: | (2S,3S,4R,5S)-2-(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)-5-[(propylsulfanyl)methyl]pyrrolidine-3,4-diol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Cameron, S.A, Firestone, R.S, Schramm, V.L, Almo, S.C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | TBA
To be published
|
|
5TC7
 
 | | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with 5'-methylthiotubercidin at 1.75 angstrom | | Descriptor: | 2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-METHYLSULFANYLMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Cameron, S.A, Firestone, R.S, Schramm, V.L, Almo, S.C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | TBA
To be published
|
|
6O1T
 
 | | BOVINE SALIVARY PROTEIN FORM 30B WITH OLEIC ACID | | Descriptor: | CALCIUM ION, OLEIC ACID, Short palate, ... | | Authors: | Zhang, H, Arcus, V.L. | | Deposit date: | 2019-02-21 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three dimensional structure of Bovine Salivary Protein 30b (BSP30b) and its interaction with specific rumen bacteria.
Plos One, 14, 2019
|
|
3D0O
 
 | |
3D4P
 
 | |
3DHD
 
 | | Crystal structure of human NAMPT complexed with nicotinamide mononucleotide and pyrophosphate | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, MAGNESIUM ION, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Ho, M, Burgos, E.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2008-06-17 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A phosphoenzyme mimic, overlapping catalytic sites and reaction coordinate motion for human NAMPT.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1YGP
 
 | | PHOSPHORYLATED FORM OF YEAST GLYCOGEN PHOSPHORYLASE WITH PHOSPHATE BOUND IN THE ACTIVE SITE. | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, YEAST GLYCOGEN PHOSPHORYLASE | | Authors: | Lin, K, Rath, V.L, Dai, S.C, Fletterick, R.J, Hwang, P.K. | | Deposit date: | 1996-05-30 | | Release date: | 1996-12-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A protein phosphorylation switch at the conserved allosteric site in GP.
Science, 273, 1996
|
|
4KEN
 
 | | Crystal Structure of AmpC beta-lactamase N152G Mutant in Complex with Cefoxitin | | Descriptor: | (2R)-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Docter, B.E, Baggett, V.L, Powers, R.A, Wallar, B.J. | | Deposit date: | 2013-04-25 | | Release date: | 2014-10-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Complexed structures of AmpC beta-lactamase
To be Published
|
|
4KG5
 
 | | Crystal Structure of AmpC beta-lactamase N152G Mutant in Complex with Cefotaxime | | Descriptor: | Beta-lactamase, CEFOTAXIME, C3' cleaved, ... | | Authors: | Docter, B.E, Baggett, V.L, Powers, R.A, Wallar, B.J. | | Deposit date: | 2013-04-28 | | Release date: | 2014-10-29 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Complexed structures of AmpC beta-lactamase
To be Published
|
|
1SQT
 
 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 7-METHOXY-8-[1-(METHYLSULFONYL)-1H-PYRAZOL-4-YL]NAPHTHALENE-2-CARBOXIMIDAMIDE, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Geyer, A, McClellan, W.J, Rockway, T.W, Weitzberg, M, Zhang, X, Mantei, R, Stewart, K, Nienaber, V, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2004-03-19 | | Release date: | 2004-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interaction with the S1beta-pocket of urokinase: 8-heterocycle substituted and 6,8-disubstituted 2-naphthamidine urokinase inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1SQO
 
 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 8-(PYRIMIDIN-2-YLAMINO)NAPHTHALENE-2-CARBOXIMIDAMIDE, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Geyer, A, McClellan, W.J, Rockway, T.W, Weitzberg, M, Zhao, X, Mantei, R, Stewart, K, Nienaber, V, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2004-03-19 | | Release date: | 2004-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Interaction with the S1beta-pocket of urokinase: 8-heterocycle substituted and 6,8-disubstituted 2-naphthamidine urokinase inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
4KG6
 
 | |
1SFH
 
 | | Reduced state of amicyanin mutant P94F | | Descriptor: | Amicyanin, COPPER (I) ION, SODIUM ION | | Authors: | Carrell, C.J, Sun, D, Jiang, S, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2004-02-19 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural Studies of Two Mutants of Amicyanin from Paracoccus denitrificans That Stabilize the Reduced State of the Copper.
Biochemistry, 43, 2004
|
|
1SF3
 
 | | Structure of the reduced form of the P94A mutant of amicyanin | | Descriptor: | Amicyanin, COPPER (I) ION, PHOSPHATE ION | | Authors: | Carrell, C.J, Sun, D, Jiang, S, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2004-02-19 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural Studies of Two Mutants of Amicyanin from Paracoccus denitrificans That Stabilize the Reduced State of the Copper.
Biochemistry, 43, 2004
|
|
4KG2
 
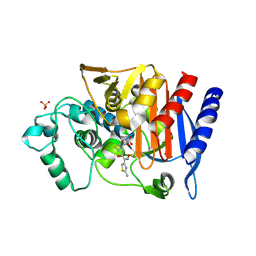 | | Crystal Structure of AmpC beta-lactamase from E. coli in Complex with Cefotaxime | | Descriptor: | Beta-lactamase, CEFOTAXIME, C3' cleaved, ... | | Authors: | Docter, B.E, Baggett, V.L, Powers, R.A, Wallar, B.J. | | Deposit date: | 2013-04-28 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Complexed structures of AmpC beta-lactamase
To be Published
|
|
1SQA
 
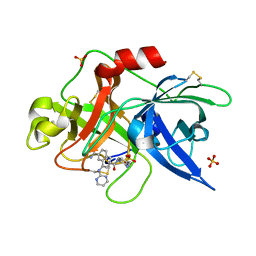 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 6-[(Z)-AMINO(IMINO)METHYL]-N-[4-(AMINOMETHYL)PHENYL]-4-(PYRIMIDIN-2-YLAMINO)-2-NAPHTHAMIDE, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Geyer, A, McClellan, W.J, Rockway, T.W, Weitzberg, M, Zhao, X, Stewart, K, Nienaber, V, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2004-03-18 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interaction with the S1beta-pocket of urokinase: 8-heterocycle substituted and 6,8-disubstituted 2-naphthamidine urokinase inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1SJ5
 
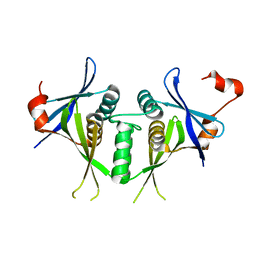 | | Crystal structure of a duf151 family protein (tm0160) from thermotoga maritima at 2.8 A resolution | | Descriptor: | conserved hypothetical protein TM0160 | | Authors: | Spraggon, G, Panatazatos, D, Klock, H.E, Wilson, I.A, Woods Jr, V.L, Lesley, S.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-03-02 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | On the use of DXMS to produce more crystallizable proteins: structures of the T. maritima proteins TM0160 and TM1171.
Protein Sci., 13, 2004
|
|
5WD6
 
 | | bovine salivary protein form 30b | | Descriptor: | CALCIUM ION, Short palate, lung and nasal epithelium carcinoma-associated protein 2B | | Authors: | Zhang, H, Arcus, V.L. | | Deposit date: | 2017-07-04 | | Release date: | 2018-07-11 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The three dimensional structure of Bovine Salivary Protein 30b (BSP30b) and its interaction with specific rumen bacteria.
Plos One, 14, 2019
|
|
5WCZ
 
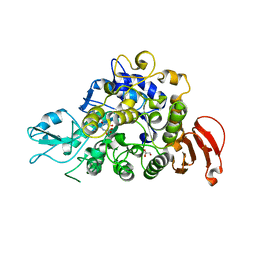 | |
