4YT9
 
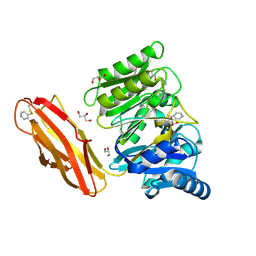 | | Crystal structure of Porphyromonas gingivalis peptidylarginine deiminase (PPAD) substrate-unbound. | | Descriptor: | GLYCEROL, Peptidylarginine deiminase, SODIUM ION | | Authors: | Goulas, T, Mizgalska, D, Garcia-Ferrer, I, Kantyka, T, Guevara, T, Szmigielski, B, Sroka, A, Millan, C, Uson, I, Veillard, F, Potempa, B, Mydel, P, Sola, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2015-03-17 | | Release date: | 2015-07-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase.
Sci Rep, 5, 2015
|
|
4YTB
 
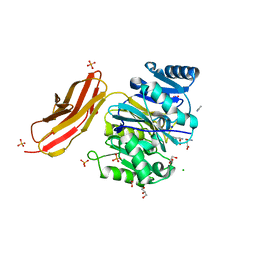 | | Crystal structure of Porphyromonas gingivalis peptidylarginine deiminase (PPAD) in complex with dipeptide Asp-Gln. | | Descriptor: | ASPARTIC ACID, AZIDE ION, CHLORIDE ION, ... | | Authors: | Goulas, T, Mizgalska, D, Garcia-Ferrer, I, Kantyka, T, Guevara, T, Szmigielski, B, Sroka, A, Millan, C, Uson, I, Veillard, F, Potempa, B, Mydel, P, Sola, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2015-03-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase.
Sci Rep, 5, 2015
|
|
5EZU
 
 | | Crystal structure of the N-terminal domain of vaccinia virus immunomodulator A46 in complex with myristic acid. | | Descriptor: | MYRISTIC ACID, Protein A46 | | Authors: | Fedosyuk, S, Bezerra, G.A, Sammito, M, Uson, I, Skern, T. | | Deposit date: | 2015-11-26 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Vaccinia Virus Immunomodulator A46: A Lipid and Protein-Binding Scaffold for Sequestering Host TIR-Domain Proteins.
PLoS Pathog., 12, 2016
|
|
4WXB
 
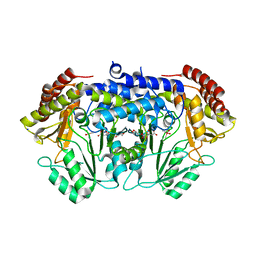 | | Crystal Structure of Serine Hydroxymethyltransferase from Streptococcus thermophilus | | Descriptor: | CACODYLATE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Hernandez, K, Zelen, I, Petrillo, G, Uson, I, Wandtke, C, Bujons, J, Joglar, J, Parella, T, Clapes, P. | | Deposit date: | 2014-11-13 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Engineered L-Serine Hydroxymethyltransferase from Streptococcus thermophilus for the Synthesis of alpha , alpha-Dialkyl-alpha-Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4WXG
 
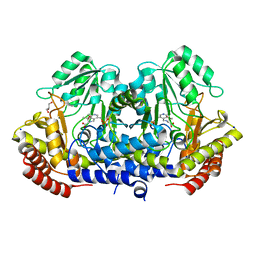 | | Crystal structure of L-Serine Hydroxymethyltransferase in complex with a mixture of L-Threonine and Glycine | | Descriptor: | GLYCEROL, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-threonine, SODIUM ION, ... | | Authors: | Hernandez, K, Zelen, I, Petrillo, G, Uson, I, Wandtke, C, Bujons, J, Joglar, J, Parella, T, Clapes, P. | | Deposit date: | 2014-11-13 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineered L-Serine Hydroxymethyltransferase from Streptococcus thermophilus for the Synthesis of alpha , alpha-Dialkyl-alpha-Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4WXF
 
 | | Crystal structure of L-Serine Hydroxymethyltransferase in complex with glycine | | Descriptor: | GLYCEROL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], Serine hydroxymethyltransferase | | Authors: | Hernandez, K, Zelen, I, Petrillo, G, Uson, I, Wadtke, C, Bujons, J, Joglar, J, Parella, T, Clapes, P. | | Deposit date: | 2014-11-13 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Engineered L-Serine Hydroxymethyltransferase from Streptococcus thermophilus for the Synthesis of alpha , alpha-Dialkyl-alpha-Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2Y8P
 
 | | Crystal Structure of an Outer Membrane-Anchored Endolytic Peptidoglycan Lytic Transglycosylase (MltE) from Escherichia coli | | Descriptor: | ENDO-TYPE MEMBRANE-BOUND LYTIC MUREIN TRANSGLYCOSYLASE A | | Authors: | Artola-Recolons, C, Carrasco-Lopez, C, Llarrull, L.I, Kumarasiri, M, Lastochkin, E, Martinez-Ilarduya, I, Meindl, K, Uson, I, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2011-02-08 | | Release date: | 2011-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | High-Resolution Crystal Structure of Mlte, an Outer Membrane-Anchored Endolytic Peptidoglycan Lytic Transglycosylase from Escherichia Coli.
Biochemistry, 50, 2011
|
|
6WCU
 
 | | Crystal structure of coiled coil region of human septin 5 | | Descriptor: | Septin-5 | | Authors: | Cabrejos, D.A.L, Cavini, I, Sala, F.A, Valadares, N.F, Pereira, H.M, Brandao-Neto, J, Nascimento, A.F.Z, Uson, I, Araujo, A.P.U, Garratt, R.C. | | Deposit date: | 2020-03-31 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Orientational Ambiguity in Septin Coiled Coils and its Structural Basis.
J.Mol.Biol., 433, 2021
|
|
6WSM
 
 | | Crystal structure of coiled coil region of human septin 8 | | Descriptor: | SULFATE ION, Septin-8 | | Authors: | Cabrejos, D.A.L, Cavini, I, Sala, F.A, Valadares, N.F, Pereira, H.M, Brandao-Neto, J, Nascimento, A.F.Z, Uson, I, Araujo, A.P.U, Garratt, R.C. | | Deposit date: | 2020-05-01 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Orientational Ambiguity in Septin Coiled Coils and its Structural Basis.
J.Mol.Biol., 433, 2021
|
|
6OR0
 
 | | Crystal structure of Insulin from Non-merohedrally twinned crystals | | Descriptor: | Insulin chain A, Insulin chain B | | Authors: | Sevvana, M, Ruf, M, Uson, I, Sheldrick, G.M, Herbst-Irmer, R. | | Deposit date: | 2019-04-29 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Non-merohedral twinning: from minerals to proteins.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6OQZ
 
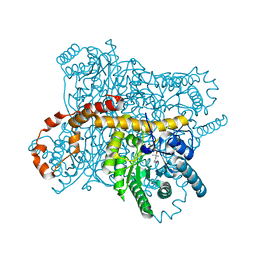 | | Crystal structure of Glucose Isomerase from Non-merohedrally twinned crystals | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Sevvana, M, Ruf, M, Uson, I, Sheldrick, G.M, Herbst-Irmer, R. | | Deposit date: | 2019-04-29 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Non-merohedral twinning: from minerals to proteins.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1VZ5
 
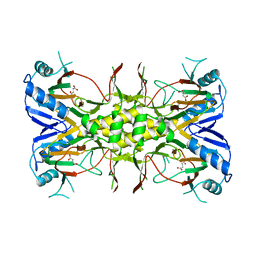 | | Succinate Complex of AtsK | | Descriptor: | PUTATIVE ALKYLSULFATASE ATSK, SUCCINIC ACID | | Authors: | Mueller, I, Stueckl, A.C, Uson, I, Kertesz, M. | | Deposit date: | 2004-05-14 | | Release date: | 2004-11-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Succinate Complex Crystal Structures of the Alpha-Ketoglutarate-Dependent Dioxygenase Atsk: Steric Aspects of Enzyme Self-Hydroxylation
J.Biol.Chem., 280, 2005
|
|
1VZ4
 
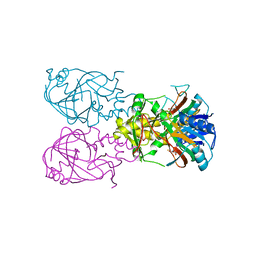 | | Fe-Succinate Complex of AtsK | | Descriptor: | FE (II) ION, PUTATIVE ALKYLSULFATASE ATSK, SUCCINIC ACID | | Authors: | Mueller, I, Stueckl, A.C, Uson, I, Kertesz, M. | | Deposit date: | 2004-05-14 | | Release date: | 2004-11-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Succinate Complex Crystal Structures of the Alpha-Ketoglutarate-Dependent Dioxygenase Atsk: Steric Aspects of Enzyme Self-Hydroxylation
J.Biol.Chem., 280, 2005
|
|
7TA2
 
 | | Crystal structure of the human sperm-expressed surface protein SPACA6 | | Descriptor: | BROMIDE ION, Sperm acrosome membrane-associated protein 6 | | Authors: | Vance, T.D.R, Yip, P, Jimenez, E, Uson, I, Lee, J.E. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SPACA6 ectodomain structure reveals a conserved superfamily of gamete fusion-associated proteins.
Commun Biol, 5, 2022
|
|
7KFL
 
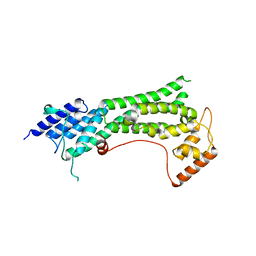 | | Crystal structure of the cargo-binding domain from the plant class XI myosin (MyoXIk) | | Descriptor: | Myosin-17 | | Authors: | Turowski, V.R, Ruiz, D.M, Nascimento, A.F.Z, Millan, C, Sammito, M.D, Juanhuix, J, Cremonesi, A.S, Uson, I, Giuseppe, P.O, Murakami, M.T. | | Deposit date: | 2020-10-14 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the class XI myosin globular tail reveals evolutionary hallmarks for cargo recognition in plants.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4GDO
 
 | | Structure of a fragment of the rod domain of plectin | | Descriptor: | Plectin | | Authors: | De Pereda, J.M, Buey, R.M, Uson, I, Sammito, M.D, De Marino, I. | | Deposit date: | 2012-08-01 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploiting tertiary structure through local folds for crystallographic phasing.
Nat.Methods, 10, 2013
|
|
3SZS
 
 | |
3FX8
 
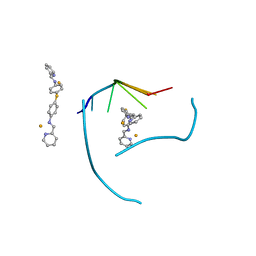 | | Distinct recognition of three-way DNA junctions by a thioester variant of a metallo-supramolecular cylinder ('helicate') | | Descriptor: | (5'-D(*CP*GP*TP*AP*CP*G)-3', 4,4'-sulfanediylbis{N-[(1E)-pyridin-2-ylmethylidene]aniline}, FE (II) ION | | Authors: | Boer, D.R, Uson, I, Coll, M. | | Deposit date: | 2009-01-20 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Self-Assembly of Functionalizable Two-Component 3D DNA Arrays through the Induced Formation of DNA Three-Way-Junction Branch Points by Supramolecular Cylinders.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3UFE
 
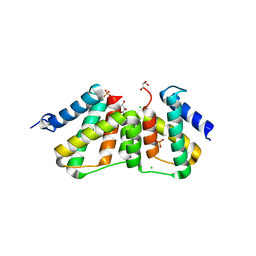 | | Structure of transcriptional antiterminator (BGLG-family) at 1.5 A resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Grosse, C, Himmel, S, Becker, S, Sheldrick, G.M, Uson, I. | | Deposit date: | 2011-11-01 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of transcriptional antiterminator (BGLG-family) at 1.5 A resolution
To be Published
|
|
5I8L
 
 | | Crystal structure of the full-length cell wall-binding module of Cpl7 mutant R223A | | Descriptor: | GLYCEROL, Lysozyme | | Authors: | Bernardo-Garcia, N, Silva-Martin, N, Uson, I, Hermoso, J.A. | | Deposit date: | 2016-02-19 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Deciphering how Cpl-7 cell wall-binding repeats recognize the bacterial peptidoglycan.
Sci Rep, 7, 2017
|
|
6I7P
 
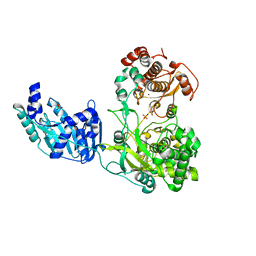 | | Crystal structure of the full-length Zika virus NS5 protein (Human isolate Z1106033) | | Descriptor: | NS5, PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Ferrero, D.S, Ruiz-Arroyo, V.M, Soler, N, Uson, I, Verdaguer, N. | | Deposit date: | 2018-11-16 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.975 Å) | | Cite: | Supramolecular arrangement of the full-length Zika virus NS5.
Plos Pathog., 15, 2019
|
|
5NDX
 
 | | The bacterial orthologue of Human a-L-iduronidase does not need N-glycan post-translational modifications to be catalytically competent: Crystallography and QM/MM insights into Mucopolysaccharidosis I | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-6-(4-methyl-2-oxidanylidene-chromen-7-yl)oxy-3,4,5-tris(oxidanyl)oxane-2-carboxylic acid, Glycosyl hydrolase, SULFATE ION | | Authors: | Raich, L, Valero-Gonzalez, J, Castro-Lopez, J, Millan, C, Jimenez-Garcia, M.J, Nieto, P, Uson, I, Hurtado-Guerrero, R, Rovira, C. | | Deposit date: | 2017-03-09 | | Release date: | 2018-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The bacterial orthologue of Human a-L-iduronidase does not need N-glycan post-translational modifications to be catalytically competent:
Crystallography and QM/MM insights into Mucopolysaccharidosis I.
To Be Published
|
|
1OIJ
 
 | | Crystal structure of the alkylsulfatase AtsK, a non-heme Fe(II) alphaketoglutarate dependent Dioxygenase in complex with alphaketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, PUTATIVE ALKYLSULFATASE ATSK, SODIUM ION | | Authors: | Mueller, I, Kahnert, A, Pape, T, Dierks, T, Meyer-Klauke, W, Kertesz, M.A, Uson, I. | | Deposit date: | 2003-06-18 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Alkylsulfatase Atsk: Insights Into the Catalytic Mechanism of the Fe(II) Alpha-Ketoglutarate-Dependent Dioxygenase Superfamily
Biochemistry, 42, 2004
|
|
1OIK
 
 | | Crystal structure of the alkylsulfatase AtsK, a non-heme Fe(II) alphaketoglutarate dependent Dioxygenase in complex with fe, alphaketoglutarate and 2-ethyl-1-hexanesulfuric acid | | Descriptor: | (2R)-2-ETHYL-1-HEXANESULFONIC ACID, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Mueller, I, Kahnert, A, Pape, T, Dierks, T, Meyer-Klauke, W, Kertesz, M.A, Uson, I. | | Deposit date: | 2003-06-18 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of the Alkylsulfatase Atsk: Insights Into the Catalytic Mechanism of the Fe(II) Alpha-Ketoglutarate-Dependent Dioxygenase Superfamily
Biochemistry, 42, 2004
|
|
1OII
 
 | | Crystal structure of the alkylsulfatase AtsK, a non-heme Fe(II) alphaketoglutarate dependent Dioxygenase in complex with iron and alphaketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, PUTATIVE ALKYLSULFATASE ATSK | | Authors: | Mueller, I, Kahnert, A, Pape, T, Dierks, T, Meyer-Klauke, W, Kertesz, M.A, Uson, I. | | Deposit date: | 2003-06-18 | | Release date: | 2004-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of the Alkylsulfatase Atsk: Insights Into the Catalytic Mechanism of the Fe(II) Alpha-Ketoglutarate-Dependent Dioxygenase Superfamily
Biochemistry, 42, 2004
|
|
