1IQ7
 
 | | Ovotransferrin, C-Terminal Lobe, Apo Form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ovotransferrin, SULFATE ION | | Authors: | Mizutani, K, Muralidhara, B.K, Yamashita, H, Tabata, S, Mikami, B, Hirose, M. | | Deposit date: | 2001-07-06 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Anion-mediated Fe3+ release mechanism in ovotransferrin C-lobe: a structurally identified SO4(2-) binding site and its implications for the kinetic pathway.
J.Biol.Chem., 276, 2001
|
|
5AZD
 
 | | Crystal structure of thermophilic rhodopsin. | | Descriptor: | Bacteriorhodopsin | | Authors: | Mizutani, K, Hashimoto, N, Tsukamoto, T, Yamashita, K, Yamamoto, M, Sudo, Y, Murata, T. | | Deposit date: | 2015-09-30 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystallographic structure of thermophilic rhodopsin: implications for high thermal stability and optogenetic availability.
To Be Published
|
|
1GEQ
 
 | |
1V9K
 
 | | The crystal structure of the catalytic domain of pseudouridine synthase RluC from Escherichia coli | | Descriptor: | Ribosomal large subunit pseudouridine synthase C, SULFATE ION | | Authors: | Machida, Y, Mizutani, K, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the catalytic domains of pseudouridine synthases RluC and RluD from Escherichia coli
Biochemistry, 43, 2004
|
|
1IT4
 
 | | Solution structure of the prokaryotic Phospholipase A2 from Streptomyces violaceoruber | | Descriptor: | CALCIUM ION, phospholipase A2 | | Authors: | Ohtani, K, Sugiyama, M, Izuhara, M, Koike, T. | | Deposit date: | 2002-01-08 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel prokaryotic phospholipase A2. Characterization, gene cloning, and solution structure.
J.BIOL.CHEM., 277, 2002
|
|
4UX2
 
 | | Cryo-EM structure of antagonist-bound E2P gastric H,K-ATPase (SCH.E2. MgF) | | Descriptor: | POTASSIUM-TRANSPORTING ATPASE ALPHA CHAIN 1, POTASSIUM-TRANSPORTING ATPASE SUBUNIT BETA | | Authors: | Abe, K, Tani, K, Fujiyoshi, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-09-17 | | Last modified: | 2014-11-12 | | Method: | ELECTRON CRYSTALLOGRAPHY (7 Å) | | Cite: | Systematic Comparison of Molecular Conformations of H+,K+-ATPase Reveals an Important Contribution of the A-M2 Linker for the Luminal Gating.
J.Biol.Chem., 289, 2014
|
|
4UX1
 
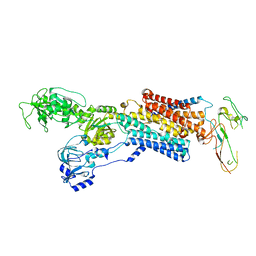 | | Cryo-EM structure of antagonist-bound E2P gastric H,K-ATPase (SCH.E2. AlF) | | Descriptor: | POTASSIUM-TRANSPORTING ATPASE ALPHA CHAIN 1, POTASSIUM-TRANSPORTING ATPASE SUBUNIT BETA | | Authors: | Abe, K, Tani, K, Fujiyoshi, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-09-17 | | Last modified: | 2014-11-12 | | Method: | ELECTRON CRYSTALLOGRAPHY (8 Å) | | Cite: | Systematic Comparison of Molecular Conformations of H+,K+-ATPase Reveals an Important Contribution of the A-M2 Linker for the Luminal Gating.
J.Biol.Chem., 289, 2014
|
|
4P79
 
 | | Crystal structure of mouse claudin-15 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Claudin-15 | | Authors: | Suzuki, H, Nishizawa, T, Tani, K, Yamazaki, Y, Tamura, A, Ishitani, R, Dohmae, N, Tsukita, S, Nureki, O, Fujiyoshi, Y. | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a claudin provides insight into the architecture of tight junctions.
Science, 344, 2014
|
|
2XZB
 
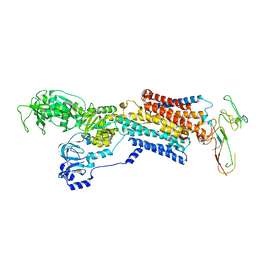 | | Pig Gastric H,K-ATPase with bound BeF and SCH28080 | | Descriptor: | POTASSIUM-TRANSPORTING ATPASE ALPHA CHAIN 1, POTASSIUM-TRANSPORTING ATPASE SUBUNIT BETA | | Authors: | Abe, K, Tani, K, Fujiyoshi, Y. | | Deposit date: | 2010-11-24 | | Release date: | 2011-01-26 | | Last modified: | 2020-09-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (7 Å) | | Cite: | Conformational Rearrangement of Gastric H(+),K(+)- ATPase Induced by an Acid Suppressant.
Nat.Commun., 2, 2011
|
|
3IXZ
 
 | | Pig gastric H+/K+-ATPase complexed with aluminium fluoride | | Descriptor: | Potassium-transporting ATPase alpha, Potassium-transporting ATPase subunit beta | | Authors: | Abe, K, Tani, K, Nishizawa, T, Fujiyoshi, Y. | | Deposit date: | 2009-03-09 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (6.5 Å) | | Cite: | Inter-subunit interaction of gastric H+,K+-ATPase prevents reverse reaction of the transport cycle
Embo J., 28, 2009
|
|
1BM1
 
 | | CRYSTAL STRUCTURE OF BACTERIORHODOPSIN IN THE LIGHT-ADAPTED STATE | | Descriptor: | BACTERIORHODOPSIN, PHOSPHORIC ACID 2,3-BIS-(3,7,11,15-TETRAMETHYL-HEXADECYLOXY)-PROPYL ESTER 2-HYDROXO-3-PHOSPHONOOXY-PROPYL ESTER, RETINAL | | Authors: | Sato, H, Takeda, K, Tani, K, Hino, T, Okada, T, Nakasako, M, Kamiya, N, Kouyama, T. | | Deposit date: | 1998-07-28 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Specific lipid-protein interactions in a novel honeycomb lattice structure of bacteriorhodopsin.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6ACF
 
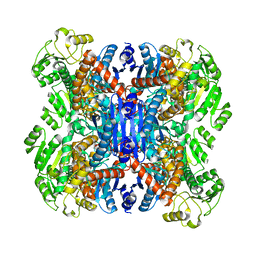 | | structure of leucine dehydrogenase from Geobacillus stearothermophilus by cryo-EM | | Descriptor: | Leucine dehydrogenase | | Authors: | Yamaguchi, H, Kamegawa, A, Nakata, K, Kashiwagi, T, Mizukoshi, T, Fujiyoshi, Y, Tani, K. | | Deposit date: | 2018-07-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into thermostabilization of leucine dehydrogenase from its atomic structure by cryo-electron microscopy
J. Struct. Biol., 205, 2019
|
|
6ACH
 
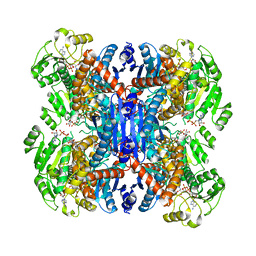 | | Structure of NAD+-bound leucine dehydrogenase from Geobacillus stearothermophilus by cryo-EM | | Descriptor: | Leucine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamaguchi, H, Kamegawa, A, Nakata, K, Kashiwagi, T, Mizukoshi, T, Fujiyoshi, Y, Tani, K. | | Deposit date: | 2018-07-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into thermostabilization of leucine dehydrogenase from its atomic structure by cryo-electron microscopy
J. Struct. Biol., 205, 2019
|
|
3IZ1
 
 | | C-alpha model fitted into the EM structure of Cx26M34A | | Descriptor: | Gap junction beta-2 protein | | Authors: | Oshima, A, Tani, K, Toloue, M.M, Hiroaki, Y, Smock, A, Inukai, S, Cone, A, Nicholson, B.J, Sosinsky, G.E, Fujiyoshi, Y. | | Deposit date: | 2010-08-19 | | Release date: | 2010-11-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (6 Å) | | Cite: | Asymmetric configurations and N-terminal rearrangements in connexin26 gap junction channels.
J.Mol.Biol., 405, 2011
|
|
3IZ2
 
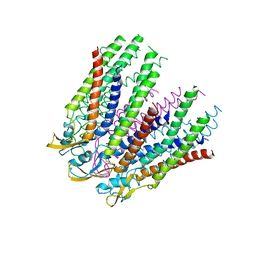 | | C-alpha model fitted into the EM structure of Cx26M34Adel2-7 | | Descriptor: | Gap junction beta-2 protein | | Authors: | Oshima, A, Tani, K, Toloue, M.M, Hiroaki, Y, Smock, A, Inukai, S, Cone, A, Nicholson, B.J, Sosinsky, G.E, Fujiyoshi, Y. | | Deposit date: | 2010-08-19 | | Release date: | 2010-11-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (10 Å) | | Cite: | Asymmetric configurations and N-terminal rearrangements in connexin26 gap junction channels.
J.Mol.Biol., 405, 2011
|
|
3IYZ
 
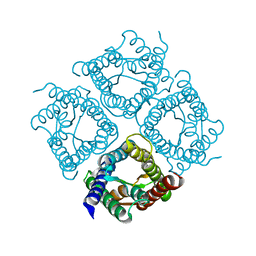 | | Structure of Aquaporin-4 S180D mutant at 10.0 A resolution from electron micrograph | | Descriptor: | Aquaporin-4 | | Authors: | Mitsuma, T, Tani, K, Hiroaki, Y, Kamegawa, A, Suzuki, H, Hibino, H, Kurachi, Y, Fujiyoshi, Y. | | Deposit date: | 2010-07-24 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (10 Å) | | Cite: | Influence of the cytoplasmic domains of aquaporin-4 on water conduction and array formation.
J.Mol.Biol., 402, 2010
|
|
7WSV
 
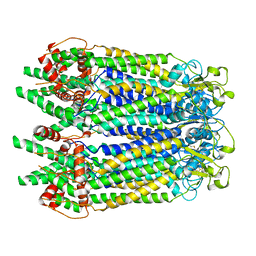 | | Cryo-EM structure of the N-terminal deletion mutant of human pannexin-1 in a nanodisc | | Descriptor: | Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2022-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
3X29
 
 | | CRYSTAL STRUCTURE of MOUSE CLAUDIN-19 IN COMPLEX with C-TERMINAL FRAGMENT OF CLOSTRIDIUM PERFRINGENS ENTEROTOXIN | | Descriptor: | Claudin-19, Heat-labile enterotoxin B chain | | Authors: | Saitoh, Y, Suzuki, H, Tani, K, Nishikawa, K, Irie, K, Ogura, Y, Tamura, A, Tsukita, S, Fujiyoshi, Y. | | Deposit date: | 2014-12-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin
Science, 347, 2015
|
|
1V9J
 
 | | Solution structure of a BolA-like protein from Mus musculus | | Descriptor: | BolA-like protein RIKEN cDNA 1110025L05 | | Authors: | Kasai, T, Inoue, M, Koshiba, S, Yabuki, T, Aoki, M, Nunokawa, E, Seki, E, Matsuda, T, Matsuda, N, Tomo, Y, Shirouzu, M, Terada, T, Obayashi, N, Hamana, H, Shinya, N, Tatsuguchi, A, Yasuda, S, Yoshida, M, Hirota, H, Matsuo, Y, Tani, K, Suzuki, H, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-26 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a BolA-like protein from Mus musculus
Protein Sci., 13, 2004
|
|
2D57
 
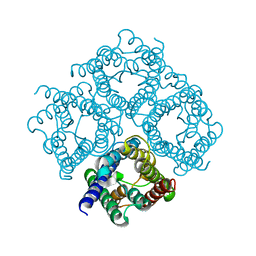 | | Double layered 2D crystal structure of AQUAPORIN-4 (AQP4M23) at 3.2 a resolution by electron crystallography | | Descriptor: | Aquaporin-4 | | Authors: | Hiroaki, Y, Tani, K, Kamegawa, A, Gyobu, N, Nishikawa, K, Suzuki, H, Walz, T, Sasaki, S, Mitsuoka, K, Kimura, K, Mizoguchi, A, Fujiyoshi, Y. | | Deposit date: | 2005-10-29 | | Release date: | 2006-01-31 | | Last modified: | 2023-11-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.2 Å) | | Cite: | Implications of the Aquaporin-4 Structure on Array Formation and Cell Adhesion
J.Mol.Biol., 355, 2005
|
|
4BGN
 
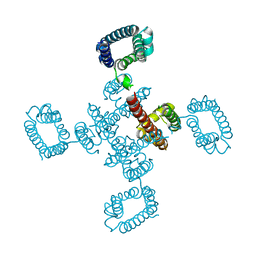 | | cryo-EM structure of the NavCt voltage-gated sodium channel | | Descriptor: | VOLTAGE-GATED SODIUM CHANNEL | | Authors: | Tsai, C.J, Tani, K, Irie, K, Hiroaki, Y, Shimomura, T, Mcmillan, D.G, Cook, G.M, Schertler, G, Fujiyoshi, Y, Li, X.D. | | Deposit date: | 2013-03-28 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | ELECTRON CRYSTALLOGRAPHY (9 Å) | | Cite: | Two Alternative Conformations of a Voltage-Gated Sodium Channel.
J.Mol.Biol., 425, 2013
|
|
5X93
 
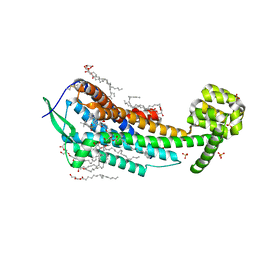 | | Human endothelin receptor type-B in complex with antagonist K-8794 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[6-[(4-tert-butylphenyl)sulfonylamino]-5-(2-methoxyphenoxy)-2-pyrimidin-2-yl-pyrimidin-4-yl]oxy-N-(2,6-dimethylphenyl)propanamide, CHOLESTEROL, ... | | Authors: | Shihoya, W, Nishizawa, T, Yamashita, K, Hirata, K, Okuta, A, Tani, K, Fujiyoshi, Y, Doi, T, Nureki, O. | | Deposit date: | 2017-03-05 | | Release date: | 2017-08-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures of endothelin ETB receptor bound to clinical antagonist bosentan and its analog
Nat. Struct. Mol. Biol., 24, 2017
|
|
5XPR
 
 | | Human endothelin receptor type-B in complex with antagonist bosentan | | Descriptor: | 4-tert-butyl-N-[6-(2-hydroxyethyloxy)-5-(2-methoxyphenoxy)-2-pyrimidin-2-yl-pyrimidin-4-yl]benzenesulfonamide, Endothelin B receptor,Endolysin,Endothelin B receptor, SULFATE ION | | Authors: | Shihoya, W, Nishizawa, T, Yamashita, K, Hirata, K, Okuta, A, Tani, K, Fujiyoshi, Y, Doi, T, Nureki, O. | | Deposit date: | 2017-06-04 | | Release date: | 2017-08-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | X-ray structures of endothelin ETB receptor bound to clinical antagonist bosentan and its analog
Nat. Struct. Mol. Biol., 24, 2017
|
|
5Y0B
 
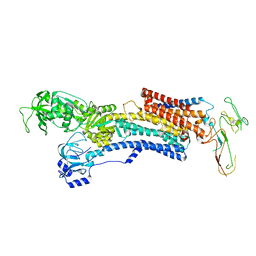 | | PIG GASTRIC H+,K+ - ATPASE IN COMPLEX with BYK99 | | Descriptor: | Potassium-transporting ATPase alpha chain 1, Potassium-transporting ATPase subunit beta | | Authors: | Abe, K, Shimokawa, J, Natio, M, Munson, K, Vagin, O, Sachs, G, Suzuki, H, Tani, K, Fujiyoshi, Y. | | Deposit date: | 2017-07-16 | | Release date: | 2017-08-09 | | Method: | ELECTRON CRYSTALLOGRAPHY (6.7 Å) | | Cite: | The cryo-EM structure of gastric H(+),K(+)-ATPase with bound BYK99, a high-affinity member of K(+)-competitive, imidazo[1,2-a]pyridine inhibitors
Sci Rep, 7, 2017
|
|
6KFG
 
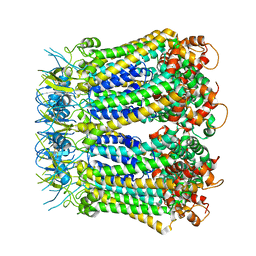 | | Undocked INX-6 hemichannel in detergent | | Descriptor: | Innexin-6 | | Authors: | Burendei, B, Shinozaki, R, Watanabe, M, Terada, T, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2019-07-07 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of undocked innexin-6 hemichannels in phospholipids.
Sci Adv, 6, 2020
|
|
