6SMY
 
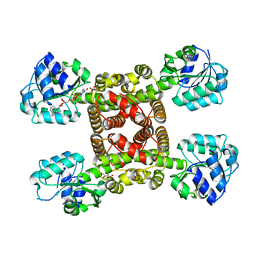 | |
6SMZ
 
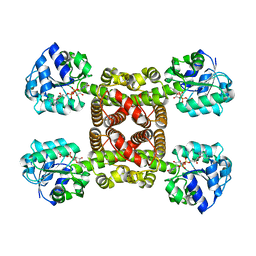 | |
6SM7
 
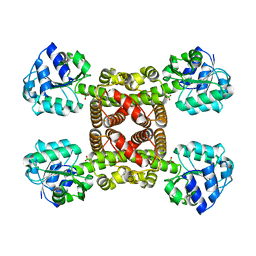 | | Crystal structure of SLA Reductase YihU from E. Coli | | Descriptor: | 3-sulfolactaldehyde reductase, BORIC ACID | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2019-08-21 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Dynamic Structural Changes Accompany the Production of Dihydroxypropanesulfonate by Sulfolactaldehyde Reductase
Acs Catalysis, 2020
|
|
6H7P
 
 | | Reductive Aminase from Aspergillus terreus in complex with NADPH4, cyclohexanone and allyl amine | | Descriptor: | CYCLOHEXANONE, Reductive Aminase, [[(2~{R},3~{S},4~{R},5~{R})-5-[(3~{R})-3-aminocarbonylpiperidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methyl hydrogen phosphate, ... | | Authors: | Sharma, M, Grogan, G, Mangas-Sanchez, J, Turner, N.J. | | Deposit date: | 2018-07-31 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of Reductive Aminase from Aspergillus terreus
Acs Catalysis, 2018
|
|
2L0J
 
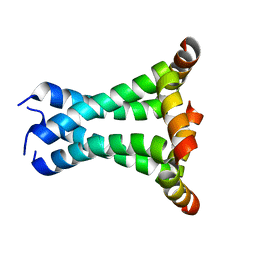 | | Solid State NMR structure of the M2 proton channel from Influenza A Virus in hydrated lipid bilayer | | Descriptor: | Matrix protein 2 | | Authors: | Sharma, M, Yi, M, Dong, H, Qin, H, Peterson, E, Busath, D.D, Zhou, H.X, Cross, T.A. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Insight into the mechanism of the influenza a proton channel from a structure in a lipid bilayer.
Science, 330, 2010
|
|
7OFY
 
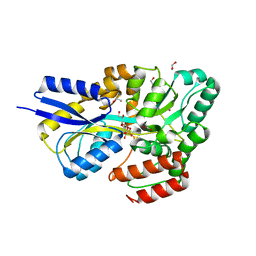 | | Crystal structure of SQ binding protein from Agrobacterium tumefaciens in complex with sulfoquinovosyl glycerol (SQGro) | | Descriptor: | 1,2-ETHANEDIOL, Sulfoquinovosyl binding protein, [(2S,3S,4S,5R,6S)-6-[(2R)-2,3-bis(oxidanyl)propoxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Jarva, M.A, Sharma, M, Goddard-Borger, E.D, Davies, G.J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oxidative desulfurization pathway for complete catabolism of sulfoquinovose by bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6K5P
 
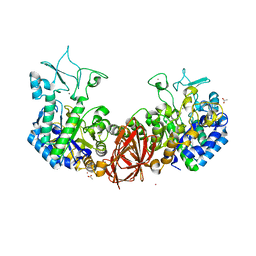 | | Structure of mosquito-larvicidal Binary toxin receptor, Cqm1 | | Descriptor: | ACETATE ION, Binary toxin receptor protein, CADMIUM ION, ... | | Authors: | Kumar, V, Sharma, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Crystal structure of BinAB toxin receptor (Cqm1) protein and molecular dynamics simulations reveal the role of unique Ca(II) ion.
Int.J.Biol.Macromol., 140, 2019
|
|
8C54
 
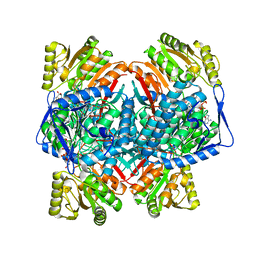 | | Cryo-EM structure of NADH bound SLA dehydrogenase RlGabD from Rhizobium leguminosarum bv. trifolii SRD1565 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Succinate semialdehyde dehydrogenase | | Authors: | Sharma, M, Meek, R.W, Armstrong, Z, Blaza, J.N, Alhifthi, A, Li, J, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2023-01-06 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Molecular basis of sulfolactate synthesis by sulfolactaldehyde dehydrogenase from Rhizobium leguminosarum.
Chem Sci, 14, 2023
|
|
2A7T
 
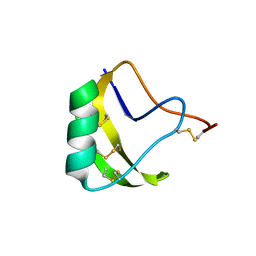 | | Crystal Structure of a novel neurotoxin from Buthus tamalus at 2.2A resolution. | | Descriptor: | Neurotoxin | | Authors: | Ethayathulla, A.S, Sharma, M, Saravanan, K, Sharma, S, Kaur, P, Yadav, S, Srinivasan, A, Singh, T.P. | | Deposit date: | 2005-07-06 | | Release date: | 2005-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a highly acidic neurotoxin from scorpion Buthus tamulus at 2.2A resolution reveals novel structural features.
J.Struct.Biol., 155, 2006
|
|
8QC3
 
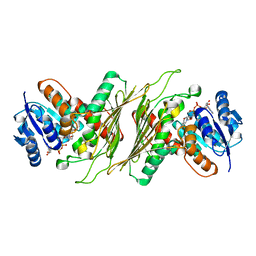 | |
8QC5
 
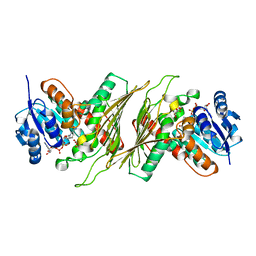 | |
8QC6
 
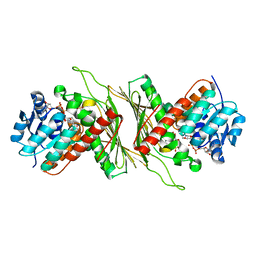 | |
7NBZ
 
 | |
7BC0
 
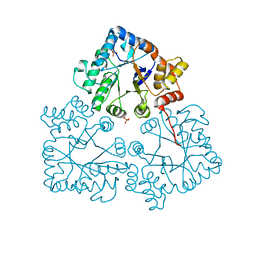 | |
7BBZ
 
 | |
7BBY
 
 | |
7BC1
 
 | |
5OCM
 
 | | Imine Reductase from Streptosporangium roseum in complex with NADP+ and 2,2,2-trifluoroacetophenone hydrate | | Descriptor: | 2,2,2-trifluoromethyl acetophenone hydrate, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sharma, M, Nestl, B, Grogan, G. | | Deposit date: | 2017-07-03 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | New imine-reducing enzymes from beta-hydroxyacid dehydrogenases by single amino acid substitutions.
Protein Eng. Des. Sel., 31, 2018
|
|
7X1X
 
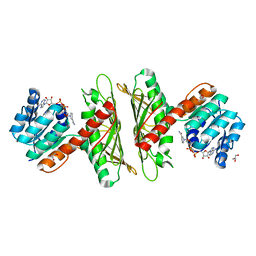 | | Crystal Structure of cis-4,5-dihydrodiol phthalate dehydrogenase in complex with NAD+ | | Descriptor: | 4,5-dihydroxyphthalate dehydrogenase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sharma, M, Mahto, J.K, Kumar, P. | | Deposit date: | 2022-02-24 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Conformational flexibility enables catalysis of phthalate cis-4,5-dihydrodiol dehydrogenase.
Arch.Biochem.Biophys., 727, 2022
|
|
7WZD
 
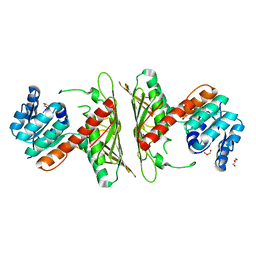 | | Crystal Structure of cis-4,5-dihydrodiol phthalate dehydrogenase from Comamonas testosteroni KF1 | | Descriptor: | 4,5-dihydroxyphthalate dehydrogenase, GLYCEROL | | Authors: | Sharma, M, Mahto, J.K, Kumar, P. | | Deposit date: | 2022-02-17 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational flexibility enables catalysis of phthalate cis-4,5-dihydrodiol dehydrogenase.
Arch.Biochem.Biophys., 727, 2022
|
|
7X2Y
 
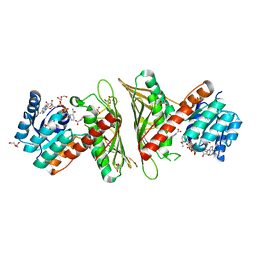 | | Crystal Structure of cis-4,5-dihydrodiol phthalate dehydrogenase in complex with NAD+ and 3-Hydroxybenzoate | | Descriptor: | 3-HYDROXYBENZOIC ACID, 4,5-dihydroxyphthalate dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sharma, M, Mahto, J.K, Kumar, P. | | Deposit date: | 2022-02-26 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Conformational flexibility enables catalysis of phthalate cis-4,5-dihydrodiol dehydrogenase.
Arch.Biochem.Biophys., 727, 2022
|
|
5OJL
 
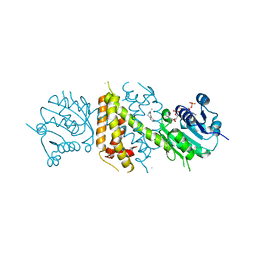 | | Imine Reductase from Aspergillus terreus in complex with NADPH4 and dibenz[c,e]azepine | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, 5-methyl-7~{H}-benzo[d][2]benzazepine, Imine reductase | | Authors: | Sharma, M, Grogan, G. | | Deposit date: | 2017-07-21 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Biocatalytic Routes to Enantiomerically Enriched Dibenz[c,e]azepines.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6SLE
 
 | | Structure of Reductive Aminase from Neosartorya fumigata in complex with NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase, putative | | Authors: | Sharma, M, Mangas-Sanchez, J, Turner, N.J, Grogan, G. | | Deposit date: | 2019-08-19 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Asymmetric synthesis of primary amines catalyzed by thermotolerant fungal reductive aminases.
Chem Sci, 11, 2020
|
|
6SKX
 
 | | Structure of Reductive Aminase from Neosartorya fumigata | | Descriptor: | Oxidoreductase, putative | | Authors: | Sharma, M, Mangas-Sanchez, J, Turner, N.J, Grogan, G. | | Deposit date: | 2019-08-16 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Asymmetric synthesis of primary amines catalyzed by thermotolerant fungal reductive aminases.
Chem Sci, 11, 2020
|
|
6TOE
 
 | |
