6RA6
 
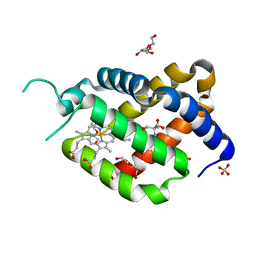 | | Ferric murine neuroglobin Gly-loop44-47/F106A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Exertier, C, Freda, I, Montemiglio, L.C, Savino, C, Vallone, B. | | Deposit date: | 2019-04-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lack of orientation selectivity of the heme insertion in murine neuroglobin revealed by resonance Raman spectroscopy.
Febs J., 287, 2020
|
|
7OHD
 
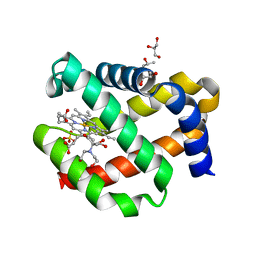 | | CRYSTAL STRUCTURE OF FERRIC MURINE NEUROGLOBIN CDLESS MUTANT | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Exertier, C, Freda, I, Montemiglio, L.C, Savino, C, Cerutti, G, Gugole, E, Vallone, B. | | Deposit date: | 2021-05-10 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the Role of Murine Neuroglobin CDloop-D-Helix Unit in CO Ligand Binding and Structural Dynamics.
Acs Chem.Biol., 17, 2022
|
|
8QRD
 
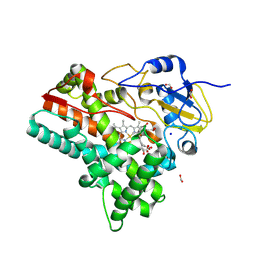 | | OleP in complex with testosterone in high salt crystallization conditions | | Descriptor: | Cytochrome P-450, FORMIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fata, F, Costanzo, A, Freda, I, Gugole, E, Bulfaro, G, Barbizzi, L, Di Renzo, M, Savino, C, Vallone, B, Montemiglio, L.C. | | Deposit date: | 2023-10-06 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | OleP in complex with testosterone in high salt crystallization conditions
To Be Published
|
|
8QYI
 
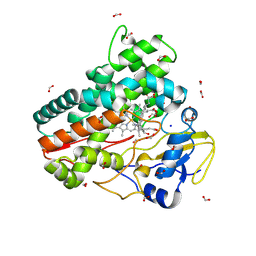 | | OleP in complex with lithocholic acid in high salt crystallization conditions | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, Cytochrome P-450, FORMIC ACID, ... | | Authors: | Fata, F, Costanzo, A, Freda, I, Gugole, E, Bulfaro, G, Barbizzi, L, Di Renzo, M, Savino, C, Vallone, B, Montemiglio, L.C. | | Deposit date: | 2023-10-26 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | OleP in complex with lithocolic acid in high salt crystallization conditions
To Be Published
|
|
1T1N
 
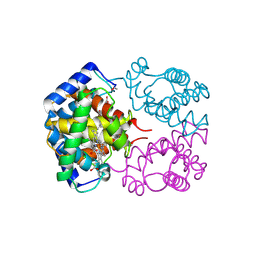 | | CRYSTAL STRUCTURE OF CARBONMONOXY HEMOGLOBIN | | Descriptor: | CARBON MONOXIDE, PROTEIN (HEMOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mazzarella, L, Vitagliano, L, Savino, C, Zagari, A. | | Deposit date: | 1999-03-05 | | Release date: | 1999-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Trematomus newnesi haemoglobin re-opens the root effect question.
J.Mol.Biol., 287, 1999
|
|
3HW7
 
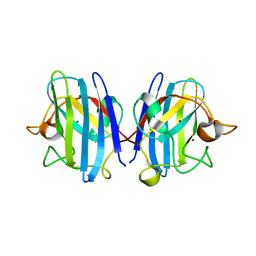 | | High pressure (0.57 GPa) crystal structure of bovine copper, zinc superoxide dismutase at 2.0 angstroms | | Descriptor: | COPPER (I) ION, COPPER (II) ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Ascone, I, Savino, C. | | Deposit date: | 2009-06-17 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexibility of the Cu,Zn superoxide dismutase structure investigated at 0.57 GPa
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3IF9
 
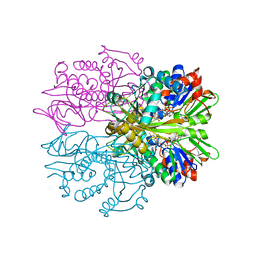 | | Crystal structure of Glycine Oxidase G51S/A54R/H244A mutant in complex with inhibitor glycolate | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCOLIC ACID, Glycine oxidase | | Authors: | Pedotti, M, Rosini, E, Molla, G, Moschetti, T, Vallone, B, Savino, C, Pollegioni, L. | | Deposit date: | 2009-07-24 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Glyphosate resistance by engineering the flavoenzyme glycine oxidase.
J.Biol.Chem., 284, 2009
|
|
1N5Q
 
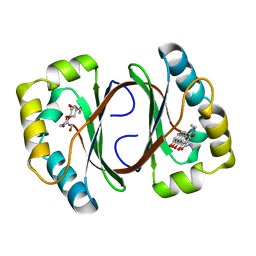 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with dehydrated Sancycline | | Descriptor: | 4-DIMETHYLAMINO-1,10,11,12-TETRAHYDROXY-3-OXO-3,4,4A,5-TETRAHYDRO-NAPHTHACENE-2-CARBOXYLIC ACID AMIDE, ActaVA-Orf6 monooxygenase, HEXAETHYLENE GLYCOL | | Authors: | Sciara, G, Kendrew, S.G, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis
Embo J., 22, 2003
|
|
1N5S
 
 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with the ligand Acetyl Dithranol | | Descriptor: | (1,8-DIHYDROXY-9-OXO-9,10-DIHYDRO-ANTHRACEN-2-YL)-ACETIC ACID, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ActVA-Orf6 monooxygenase | | Authors: | Sciara, G, Kendrew, S.G, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in
actinorhodin biosynthesis
Embo J., 22, 2003
|
|
1N5T
 
 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with the ligand Oxidized Acetyl Dithranol | | Descriptor: | (1,8-DIHYDROXY-9,10-DIOXO-9,10-DIHYDRO-ANTHRACEN-2-YL)-ACETIC ACID, ActVA-Orf6 monooxygenase | | Authors: | Sciara, G, G Kendrew, S, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in
actinorhodin biosynthesis
Embo J., 22, 2003
|
|
1N5V
 
 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with the ligand Nanaomycin D | | Descriptor: | 7-HYDROXY-5-METHYL-3,3A,5,11B-TETRAHYDRO-1,4-DIOXA-CYCLOPENTA[A]ANTHRACENE-2,6,11-TRIONE, ActVA-Orf6 monooxygenase | | Authors: | Sciara, G, Kendrew, S.G, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in
actinorhodin biosynthesis
Embo J., 22, 2003
|
|
5MNS
 
 | | Structural and functional characterization of OleP in complex with 6DEB in sodium formate | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Parisi, G, Savino, C, Montemiglio, L.C, Vallone, B. | | Deposit date: | 2016-12-13 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Substrate-induced conformational change in cytochrome P450 OleP.
FASEB J., 33, 2019
|
|
5MNV
 
 | |
4O1T
 
 | | Crystal structure of murine neuroglobin mutant F106W | | Descriptor: | Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Avella, G, Savino, C, Vallone, B. | | Deposit date: | 2013-12-16 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering the internal cavity of neuroglobin demonstrates the role of the haem-sliding mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4O2G
 
 | | Crystal structure of carbomonoxy murine neuroglobin mutant V140W | | Descriptor: | CARBON MONOXIDE, Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Avella, G, Savino, C, Vallone, B. | | Deposit date: | 2013-12-17 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Engineering the internal cavity of neuroglobin demonstrates the role of the haem-sliding mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NZI
 
 | | Crystal structure of murine neuroglobin mutant V140W | | Descriptor: | Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Avella, G, Savino, C, Vallone, B. | | Deposit date: | 2013-12-12 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering the internal cavity of neuroglobin demonstrates the role of the haem-sliding mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4O35
 
 | | Crystal structure of carbomonoxy murine neuroglobin mutant F106W | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CARBON MONOXIDE, Neuroglobin, ... | | Authors: | Avella, G, Savino, C, Vallone, B. | | Deposit date: | 2013-12-18 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering the internal cavity of neuroglobin demonstrates the role of the haem-sliding mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MU5
 
 | | Crystal structure of murine neuroglobin mutant M144W | | Descriptor: | Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Vallone, B, Avella, G, Savino, C, Ardiccioni, C, Brunori, M. | | Deposit date: | 2013-09-20 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering the internal cavity of neuroglobin demonstrates the role of the haem-sliding mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1DM1
 
 | | 2.0 A CRYSTAL STRUCTURE OF THE DOUBLE MUTANT H(E7)V, T(E10)R OF MYOGLOBIN FROM APLYSIA LIMACINA | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Federici, L, Savino, C, Musto, R, Travaglini-Allocatelli, C, Cutruzzola, F, Brunori, M. | | Deposit date: | 1999-12-13 | | Release date: | 2000-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering His(E7) affects the control of heme reactivity in Aplysia limacina myoglobin.
Biochem.Biophys.Res.Commun., 269, 2000
|
|
6H6J
 
 | | Carbomonoxy murine neuroglobin Gly-loop mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CARBON MONOXIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Exertier, C, Vallone, B, Savino, C, Freda, I, Montemiglio, L.C, Cerutti, G, Scaglione, A, Parisi, G. | | Deposit date: | 2018-07-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Proximal and distal control for ligand binding in neuroglobin: role of the CD loop and evidence for His64 gating.
Sci Rep, 9, 2019
|
|
6H5Z
 
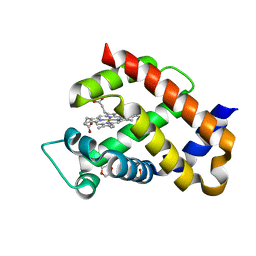 | | Ferric murine neuroglobin F106A mutant | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Exertier, C, Vallone, B, Savino, C, Freda, I, Montemiglio, L.C, Cerutti, G, Scaglione, A, Parisi, G. | | Deposit date: | 2018-07-25 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proximal and distal control for ligand binding in neuroglobin: role of the CD loop and evidence for His64 gating.
Sci Rep, 9, 2019
|
|
6H6I
 
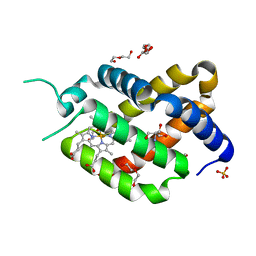 | | Ferric murine neuroglobin Gly-loop mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Exertier, C, Vallone, B, Savino, C, Freda, I, Montemiglio, L.C, Cerutti, G, Scaglione, A, Parisi, G. | | Deposit date: | 2018-07-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Proximal and distal control for ligand binding in neuroglobin: role of the CD loop and evidence for His64 gating.
Sci Rep, 9, 2019
|
|
6H6C
 
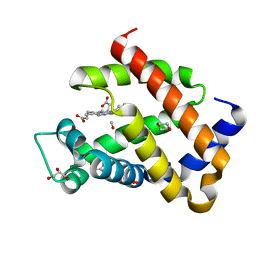 | | Carbomonoxy murine neuroglobin F106A mutant | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CARBON MONOXIDE, GLYCEROL, ... | | Authors: | Exertier, C, Vallone, B, Savino, C, Freda, I, Montemiglio, L.C, Cerutti, G, Scaglione, A, Parisi, G. | | Deposit date: | 2018-07-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Proximal and distal control for ligand binding in neuroglobin: role of the CD loop and evidence for His64 gating.
Sci Rep, 9, 2019
|
|
6H5I
 
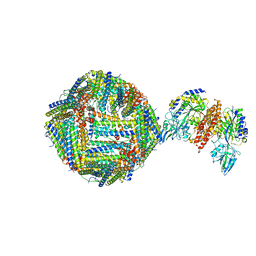 | | Single Particle Cryo-EM map of human Transferrin receptor 1 - H-Ferritin complex. | | Descriptor: | Ferritin heavy chain, Transferrin receptor protein 1 | | Authors: | Testi, C, Montemiglio, L.C, Vallone, B, Des Georges, A, Boffi, A, Mancia, F, Baiocco, P, Savino, C. | | Deposit date: | 2018-07-24 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the human ferritin-transferrin receptor 1 complex.
Nat Commun, 10, 2019
|
|
6ZI7
 
 | | Crystal structure of OleP-oleandolide(DEO) bound to L-rhamnose | | Descriptor: | (3~{R},4~{S},5~{R},6~{S},7~{S},9~{S},11~{R},12~{S},13~{R},14~{R})-3,5,7,9,11,13,14-heptamethyl-4,6,12-tris(oxidanyl)-1-oxacyclotetradecane-2,10-dione, Cytochrome P-450, FORMIC ACID, ... | | Authors: | Montemiglio, L.C, Savino, C, Vallone, B, Parisi, G, Freda, I. | | Deposit date: | 2020-06-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
