8WOS
 
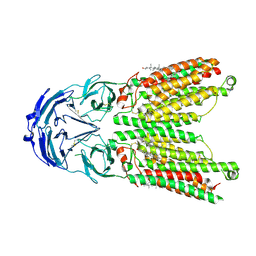 | | Cryo-EM structure of human SIDT1 protein with C1 symmetry at low pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Liu, W, Tang, M, Wang, J, Zhang, X, Wu, S, Ru, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-03-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural insights into cholesterol transport and hydrolase activity of a putative human RNA transport protein SIDT1.
Cell Discov, 10, 2024
|
|
4EF4
 
 | | Crystal structure of STING CTD complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Transmembrane protein 173 | | Authors: | Ouyang, S, Ru, H, Shaw, N, Jiang, Y, Niu, F, Zhu, Y, Qiu, W, Li, Y, Liu, Z.-J. | | Deposit date: | 2012-03-29 | | Release date: | 2012-05-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding
Immunity, 36, 2012
|
|
4EF5
 
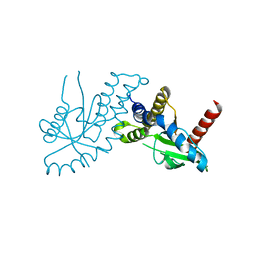 | | Crystal structure of STING CTD | | Descriptor: | Transmembrane protein 173 | | Authors: | Ouyang, S, Ru, H, Shaw, N, Jiang, Y, Niu, F, Zhu, Y, Qiu, W, Li, Y, Liu, Z.-J. | | Deposit date: | 2012-03-29 | | Release date: | 2012-05-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding
Immunity, 36, 2012
|
|
6DLP
 
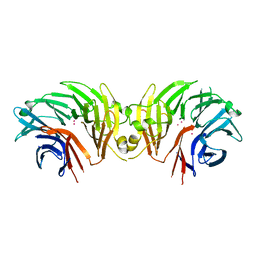 | | Crystal structure of LRRK2 WD40 domain dimer | | Descriptor: | Leucine-rich repeat serine/threonine-protein kinase 2, PLATINUM (II) ION | | Authors: | Zhang, P, Ru, H, Wang, L, Wu, H. | | Deposit date: | 2018-06-02 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structure of the WD40 domain dimer of LRRK2.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6DLO
 
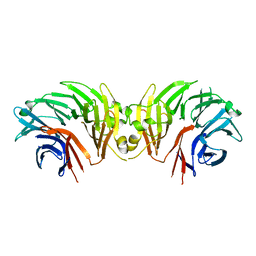 | |
9J31
 
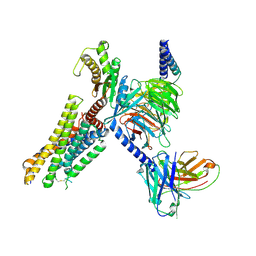 | | cryo-EM structure of zebrafish GPR4-Gs complex at pH 8.5 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Ma, Y.T, Tang, M.Y, Song, G.J, Ru, H. | | Deposit date: | 2024-08-07 | | Release date: | 2025-01-22 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cryo-EM structure of an activated GPR4-Gs signaling complex.
Nat Commun, 16, 2025
|
|
9IP3
 
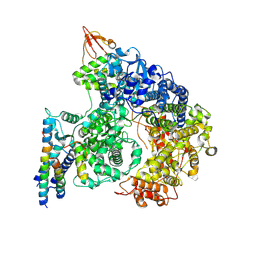 | | Cryo-EM structure of the RNA-dependent RNA polymerase complex in a compact conformation from Ebola virus | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Polymerase cofactor VP35, Maltose/maltodextrin-binding periplasmic protein,RNA-directed RNA polymerase L, ZINC ION | | Authors: | Li, G, Du, T, Wang, J, Wu, S, Ru, H. | | Deposit date: | 2024-07-10 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the RNA-dependent RNA polymerase complexes from highly pathogenic Marburg and Ebola viruses.
Nat Commun, 16, 2025
|
|
9IP4
 
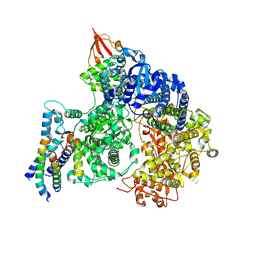 | | Cryo-EM structure of the RNA-dependent RNA polymerase complex from Marburg virus | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Polymerase cofactor VP35, RNA-directed RNA polymerase L,Maltose/maltodextrin-binding periplasmic protein, ZINC ION | | Authors: | Li, G, Du, T, Wang, J, Wu, S, Ru, H. | | Deposit date: | 2024-07-10 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural insights into the RNA-dependent RNA polymerase complexes from highly pathogenic Marburg and Ebola viruses.
Nat Commun, 16, 2025
|
|
9IP2
 
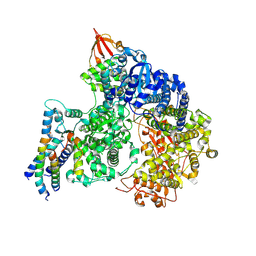 | | Cryo-EM structure of the RNA-dependent RNA polymerase complex from Marburg virus | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Polymerase cofactor VP35, RNA-directed RNA polymerase L,Maltose/maltodextrin-binding periplasmic protein, ZINC ION | | Authors: | Li, G, Du, T, Wang, J, Wu, S, Ru, H. | | Deposit date: | 2024-07-10 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the RNA-dependent RNA polymerase complexes from highly pathogenic Marburg and Ebola viruses.
Nat Commun, 16, 2025
|
|
7VYV
 
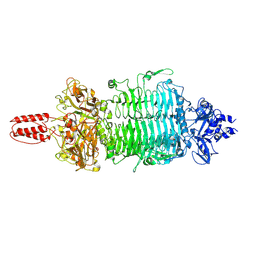 | | Cryo-EM structure of Depo32, a Klebsiella phage depolymerase targets the K2 serotype K. pneumoniae | | Descriptor: | Depolymerase | | Authors: | Cai, R, Ren, Z, Zhao, R, Wang, X, Guo, Z, Du, R, Han, W, Ru, H, Gu, J. | | Deposit date: | 2021-11-15 | | Release date: | 2023-08-30 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | Structural biology and functional features of phage-derived depolymerase Depo32 on Klebsiella pneumoniae with K2 serotype capsular polysaccharides.
Microbiol Spectr, 11, 2023
|
|
7VZ3
 
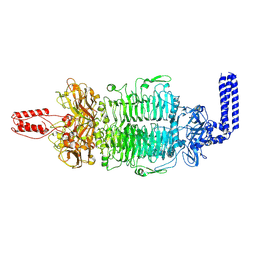 | | Cryo-EM structure of Depo32, a Klebsiella phage depolymerase targets the K2 serotype K. pneumoniae | | Descriptor: | Depolymerase | | Authors: | Cai, R, Ren, Z, Zhao, R, Wang, X, Guo, Z, Du, R, Han, W, Ru, H, Gu, J. | | Deposit date: | 2021-11-15 | | Release date: | 2023-08-30 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural biology and functional features of phage-derived depolymerase Depo32 on Klebsiella pneumoniae with K2 serotype capsular polysaccharides.
Microbiol Spectr, 11, 2023
|
|
4M4E
 
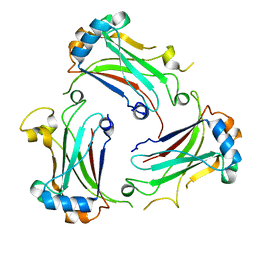 | | TRAF domain of human TRAF4 | | Descriptor: | TNF receptor-associated factor 4 | | Authors: | Niu, F, Ru, H, Ding, W, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-08-07 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural biology study of human TNF receptor associated factor 4 TRAF domain
Protein Cell, 4, 2013
|
|
4NOK
 
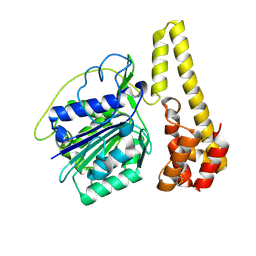 | | Crystal structure of proenzyme asparaginyl endopeptidase (AEP)/Legumain at pH 7.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
4NOM
 
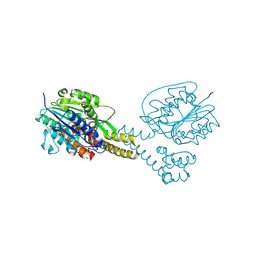 | | Crystal structure of asparaginyl endopeptidase (AEP)/Legumain activated at pH 4.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
4NOJ
 
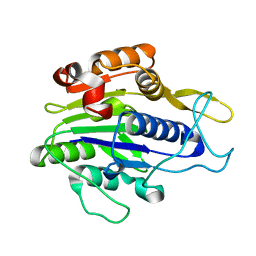 | | Crystal structure of the mature form of asparaginyl endopeptidase (AEP)/Legumain activated at pH 3.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
4NOL
 
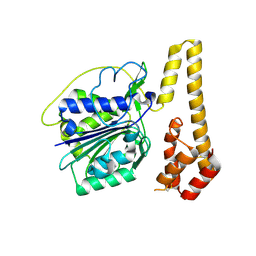 | | Crystal structure of proenzyme asparaginyl endopeptidase (AEP)/Legumain mutant D233A at pH 7.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
4QGU
 
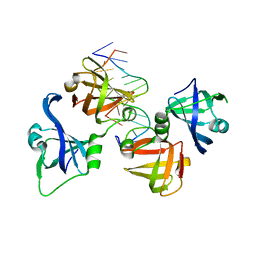 | | protein domain complex with ssDNA | | Descriptor: | DNA (5'-D(P*AP*GP*GP*CP*CP*GP*GP*CP*GP*TP*GP*A)-3'), Gamma-interferon-inducible protein 16 | | Authors: | Ni, X, Ru, H, Zhao, L, Shaw, N, Ding, W, Songying, O, Liu, Z.-J. | | Deposit date: | 2014-05-25 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | New insights into the structural basis of DNA recognition by HINa and HINb domains of IFI16.
J Mol Cell Biol, 8, 2016
|
|
8ZFZ
 
 | | cryo-EM structure of Gs-coupled zebrafish GPR4 at pH 6.5 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Ma, Y, Tang, M, Ru, H, Song, G. | | Deposit date: | 2024-05-08 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of an activated GPR4-Gs signaling complex.
Nat Commun, 16, 2025
|
|
