4M0Y
 
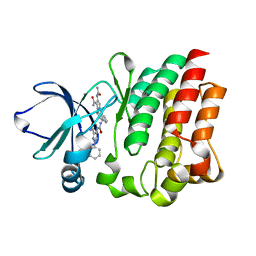 | |
8ARJ
 
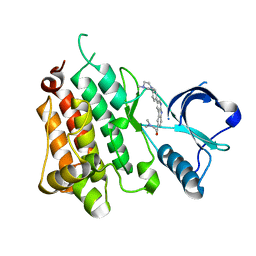 | | Anaplastic Lymphoma Kinase with a novel carboline inhibitor | | Descriptor: | 3-(dimethylamino)-1-[3-[4-(4-methylpiperazin-1-yl)phenyl]-9~{H}-pyrido[2,3-b]indol-6-yl]prop-2-en-1-one, ALK tyrosine kinase receptor | | Authors: | Mologni, L, Scapozza, L, Gambacorti-Passerini, C, Tardy, S, Goekjian, P, Robinson, C, Brown, D. | | Deposit date: | 2022-08-17 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.645 Å) | | Cite: | Discovery of Novel alpha-Carboline Inhibitors of the Anaplastic Lymphoma Kinase.
Acs Omega, 7, 2022
|
|
6N7B
 
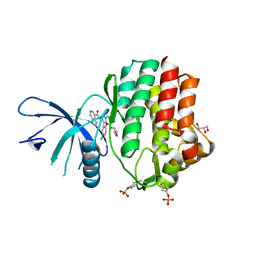 | | Structure of the human JAK1 kinase domain with compound 38 | | Descriptor: | GLYCEROL, N-[3-(5-chloro-2-methoxyphenyl)-1-methyl-1H-pyrazol-4-yl]-1H-pyrazolo[4,3-c]pyridine-7-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6N79
 
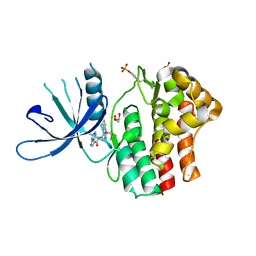 | | Structure of the human JAK1 kinase domain with compound 20 | | Descriptor: | GLYCEROL, N-{5-[5-chloro-2-(difluoromethoxy)phenyl]-1H-pyrazol-4-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6N78
 
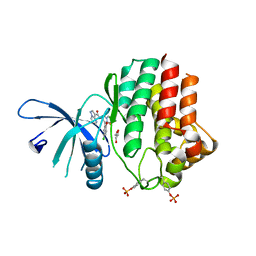 | | Structure of the human JAK1 kinase domain with compound 21 | | Descriptor: | GLYCEROL, N-{3-[5-chloro-2-(difluoromethoxy)phenyl]-1-methyl-1H-pyrazol-4-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6N7A
 
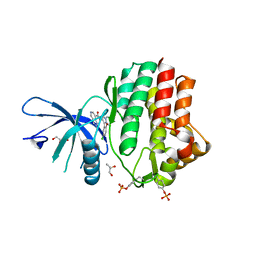 | | Structure of the human JAK1 kinase domain with compound 39 | | Descriptor: | GLYCEROL, N-[3-(5-chloro-2-methoxyphenyl)-1-methyl-1H-pyrazol-4-yl]-2-methyl-2H-pyrazolo[4,3-c]pyridine-7-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6N7C
 
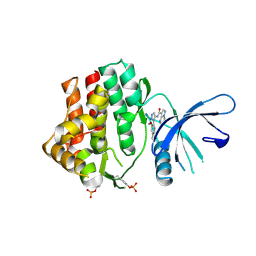 | | Structure of the human JAK1 kinase domain with compound 56 | | Descriptor: | GLYCEROL, N-[5-(3-methoxynaphthalen-2-yl)-1H-pyrazol-4-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6N77
 
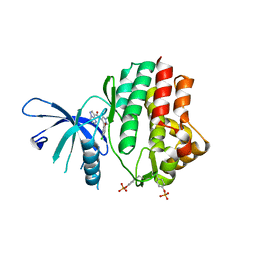 | | Structure of the human JAK1 kinase domain with compound 15 | | Descriptor: | GLYCEROL, N-[3-(5-chloro-2-methoxyphenyl)-1-methyl-1H-pyrazol-4-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6N7D
 
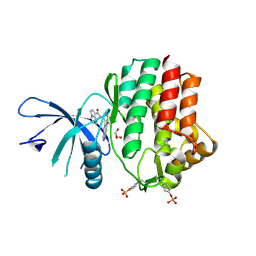 | | Structure of the human JAK1 kinase domain with compound 54 | | Descriptor: | GLYCEROL, N-[5-(6-methoxy-1H-indazol-5-yl)-1H-pyrazol-4-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
7NRF
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with darobactin (crystal form 2) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Darobactin, MAGNESIUM ION, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
7NRE
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with darobactin (crystal form 1) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Darobactin, MAGNESIUM ION, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
7NRI
 
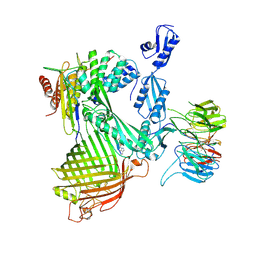 | | Structure of the darobactin-bound E. coli BAM complex (BamABCDE) | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
5JFX
 
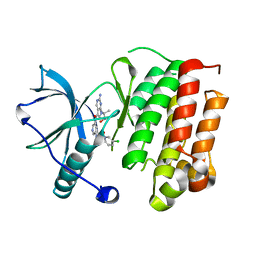 | | Crystal structure of TrkA in complex with PF-06273340 | | Descriptor: | High affinity nerve growth factor receptor, N-{5-[2-amino-7-(1-hydroxy-2-methylpropan-2-yl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonyl]pyridin-3-yl}-2-(5-chloropyridin-2-yl)acetamide | | Authors: | Jayasankar, J, Kurumbail, R, Skerratt, S, Brown, D. | | Deposit date: | 2016-04-19 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The Discovery of a Potent, Selective, and Peripherally Restricted Pan-Trk Inhibitor (PF-06273340) for the Treatment of Pain.
J. Med. Chem., 59, 2016
|
|
5JFW
 
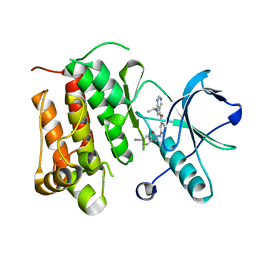 | | Crystal structure of TrkA in complex with PF-05247452 | | Descriptor: | 2-(4-cyanophenyl)-N-{5-[7-(propan-2-yl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonyl]pyridin-3-yl}acetamide, High affinity nerve growth factor receptor | | Authors: | Jayasankar, J, Kurumbail, R, Skerratt, S, Brown, D. | | Deposit date: | 2016-04-19 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The Discovery of a Potent, Selective, and Peripherally Restricted Pan-Trk Inhibitor (PF-06273340) for the Treatment of Pain.
J. Med. Chem., 59, 2016
|
|
5JFS
 
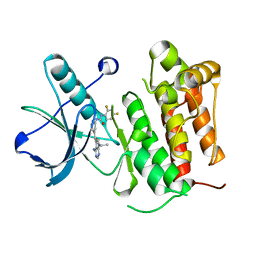 | | Crystal structure of TrkA in complex with PF-00593174 | | Descriptor: | High affinity nerve growth factor receptor, N-{4-[4-amino-7-(propan-2-yl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonyl]pyridin-2-yl}-N'-(2,4-difluorophenyl)urea | | Authors: | Jayasankar, J, Brown, D, Skerratt, S, Kurumbail, R. | | Deposit date: | 2016-04-19 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The Discovery of a Potent, Selective, and Peripherally Restricted Pan-Trk Inhibitor (PF-06273340) for the Treatment of Pain.
J. Med. Chem., 59, 2016
|
|
5JFV
 
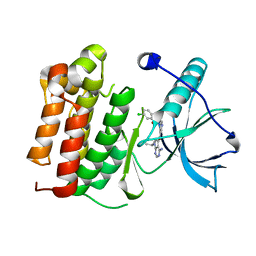 | | Crystal structure of TrkA in complex with PF-05206283 | | Descriptor: | High affinity nerve growth factor receptor, N-{5-[4-amino-7-(propan-2-yl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonyl]pyridin-3-yl}-2-(4-chlorophenyl)acetamide | | Authors: | Jayasankar, J, Kurumbail, R, Brown, D, Skerratt, S. | | Deposit date: | 2016-04-19 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The Discovery of a Potent, Selective, and Peripherally Restricted Pan-Trk Inhibitor (PF-06273340) for the Treatment of Pain.
J. Med. Chem., 59, 2016
|
|
3DEP
 
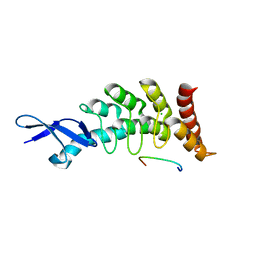 | | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43 | | Descriptor: | CHLORIDE ION, Signal recognition particle 43 kDa protein, YPGGSFDPLGLA | | Authors: | Holdermann, I, Stengel, K.F, Wild, K, Sinning, I. | | Deposit date: | 2008-06-10 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43.
Science, 321, 2008
|
|
2JL4
 
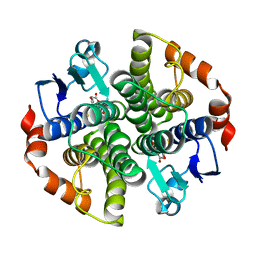 | | Holo structure of Maleyl Pyruvate Isomerase, a bacterial glutathione- s-transferase in Zeta class | | Descriptor: | GLUTATHIONE, MALEYLPYRUVATE ISOMERASE | | Authors: | Shoemark, D.K, Y Zhou, N, Williams, P.A, Hadfield, A.T. | | Deposit date: | 2008-09-04 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Bacterial Glutathione-S-Transferase Maleyl Pyruvate Isomerase and Implications for Mechanism of Isomerisation.
J.Mol.Biol., 384, 2008
|
|
2V6K
 
 | | Structure of Maleyl Pyruvate Isomerase, a bacterial glutathione-s- transferase in Zeta class, in complex with substrate analogue dicarboxyethyl glutathione | | Descriptor: | ACETATE ION, GAMMA-GLUTAMYL-S-(1,2-DICARBOXYETHYL)CYSTEINYLGLYCINE, MALEYLPYRUVATE ISOMERASE, ... | | Authors: | Shoemark, D.K, Zhou, N.-Y, Williams, P.A, Hadfield, A.T. | | Deposit date: | 2007-07-19 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of Bacterial Glutathione-S-Transferase Maleyl Pyruvate Isomerase and Implications for Mechanism of Isomerisation.
J.Mol.Biol., 384, 2008
|
|
