1ZFO
 
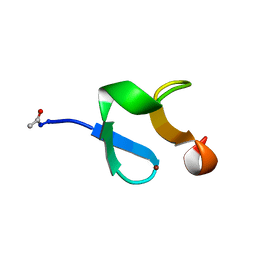 | | AMINO-TERMINAL LIM-DOMAIN PEPTIDE OF LASP-1, NMR | | Descriptor: | LASP-1, ZINC ION | | Authors: | Hammarstrom, A, Berndt, K.D, Sillard, R, Adermann, K, Otting, G. | | Deposit date: | 1996-05-06 | | Release date: | 1996-11-08 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a naturally-occurring zinc-peptide complex demonstrates that the N-terminal zinc-binding module of the Lasp-1 LIM domain is an independent folding unit.
Biochemistry, 35, 1996
|
|
2AYA
 
 | |
1J3G
 
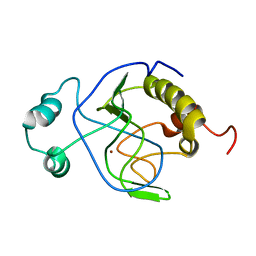 | | Solution structure of Citrobacter Freundii AmpD | | Descriptor: | AmpD protein, ZINC ION | | Authors: | Liepinsh, E, Genereux, C, Dehareng, D, Joris, B, Otting, G. | | Deposit date: | 2003-01-31 | | Release date: | 2003-02-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Citrobacter freundii AmpD, Comparison with Bacteriophage T7 Lysozyme
and Homology with PGRP Domains
J.Mol.Biol., 327, 2003
|
|
1J5B
 
 | | Solution structure of a hydrophobic analogue of the winter flounder antifreeze protein | | Descriptor: | Antifreeze protein type 1 analogue | | Authors: | Liepinsh, E, Otting, G, Harding, M.M, Ward, L.G, Mackay, J.P, Haymet, A.D. | | Deposit date: | 2002-03-22 | | Release date: | 2002-03-27 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a hydrophobic analogue of the winter flounder antifreeze protein.
Eur.J.Biochem., 269, 2002
|
|
1G7E
 
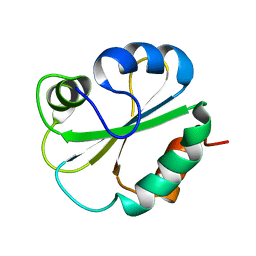 | | NMR STRUCTURE OF N-DOMAIN OF ERP29 PROTEIN | | Descriptor: | ENDOPLASMIC RETICULUM PROTEIN ERP29 | | Authors: | Liepinsh, E, Mkrtchian, S, Barishev, M, Sharipo, M, Ingelman-Sundberg, M, Otting, G. | | Deposit date: | 2000-11-10 | | Release date: | 2000-11-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Thioredoxin fold as homodimerization module in the putative chaperone ERp29: NMR structures of the domains and experimental model of the 51 kDa dimer.
Structure, 9, 2001
|
|
1JBI
 
 | | NMR structure of the LCCL domain | | Descriptor: | cochlin | | Authors: | Liepinsh, E, Trexler, M, Kaikkonen, A, Weigelt, J, Banyai, L, Patthy, L, Otting, G. | | Deposit date: | 2001-06-05 | | Release date: | 2001-10-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the LCCL domain and implications for DFNA9 deafness disorder.
EMBO J., 20, 2001
|
|
1JWE
 
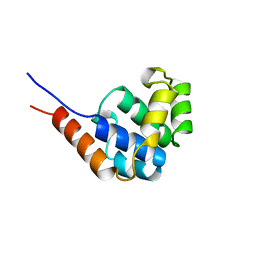 | | NMR Structure of the N-Terminal Domain of E. Coli Dnab Helicase | | Descriptor: | PROTEIN (DNAB HELICASE) | | Authors: | Weigelt, J, Brown, S.E, Miles, C.S, Dixon, N.E, Otting, G. | | Deposit date: | 1999-01-22 | | Release date: | 1999-01-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the N-terminal domain of E. coli DnaB helicase: implications for structure rearrangements in the helicase hexamer.
Structure Fold.Des., 7, 1999
|
|
2DDI
 
 | |
2DDJ
 
 | |
2HAJ
 
 | |
2D3J
 
 | |
3GRX
 
 | | NMR STRUCTURE OF ESCHERICHIA COLI GLUTAREDOXIN 3-GLUTATHIONE MIXED DISULFIDE COMPLEX, 20 STRUCTURES | | Descriptor: | GLUTAREDOXIN 3, GLUTATHIONE | | Authors: | Nordstrand, K, Aslund, F, Holmgren, A, Otting, G, Berndt, K.D. | | Deposit date: | 1998-08-17 | | Release date: | 1999-03-30 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Escherichia coli glutaredoxin 3-glutathione mixed disulfide complex: implications for the enzymatic mechanism.
J.Mol.Biol., 286, 1999
|
|
9CLD
 
 | | Crystal structure of maltose binding protein (Apo) | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Habel, E, Frkic, R.L, Jackson, C.J, Huber, T, Otting, G. | | Deposit date: | 2024-07-10 | | Release date: | 2024-12-18 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Rendering Proteins Fluorescent Inconspicuously: Genetically Encoded 4-Cyanotryptophan Conserves Their Structure and Enables the Detection of Ligand Binding Sites.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9CLC
 
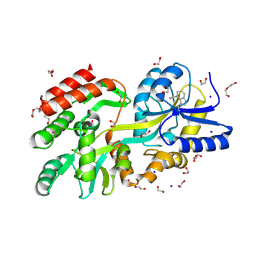 | | Crystal structure of maltose binding protein (Apo), mutant Trp10 to 4-Cyanotryptophan | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Habel, E, Frkic, R.L, Jackson, C.J, Huber, T, Otting, G. | | Deposit date: | 2024-07-10 | | Release date: | 2024-12-18 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Rendering Proteins Fluorescent Inconspicuously: Genetically Encoded 4-Cyanotryptophan Conserves Their Structure and Enables the Detection of Ligand Binding Sites.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
7RFD
 
 | |
1FTZ
 
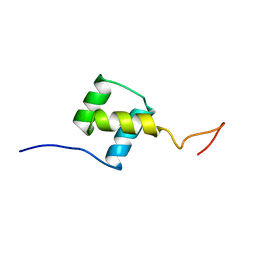 | | NUCLEAR MAGNETIC RESONANCE SOLUTION STRUCTURE OF THE FUSHI TARAZU HOMEODOMAIN FROM DROSOPHILA AND COMPARISON WITH THE ANTENNAPEDIA HOMEODOMAIN | | Descriptor: | FUSHI TARAZU PROTEIN | | Authors: | Qian, Y.Q, Furukubo-Tokunaga, K, Resendez-Perez, D, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1994-01-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the fushi tarazu homeodomain from Drosophila and comparison with the Antennapedia homeodomain.
J.Mol.Biol., 238, 1994
|
|
1SAN
 
 | |
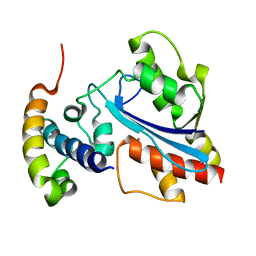
2XY8
 
 | |
7RNW
 
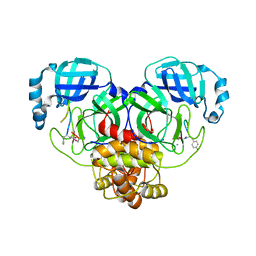 | |
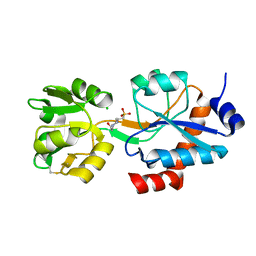
6WUP
 
 | |
4GX9
 
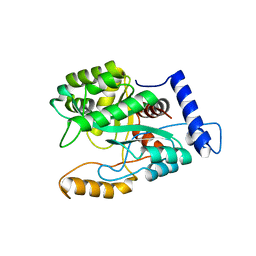 | | Crystal structure of a DNA polymerase III alpha-epsilon chimera | | Descriptor: | DNA polymerase III subunit epsilon,DNA polymerase III subunit alpha | | Authors: | Li, N, Horan, N, Xu, Z.-Q, Jacques, D, Dixon, N.E, Oakley, A.J. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits alpha, {varepsilon}, theta and beta reveals a highly flexible arrangement of the proofreading domain
Nucleic Acids Res., 41, 2013
|
|
4MZ9
 
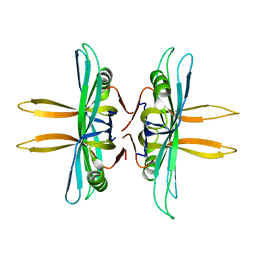 | | Revised structure of E. coli SSB | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Oakley, A.J. | | Deposit date: | 2013-09-29 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Intramolecular binding mode of the C-terminus of Escherichia coli single-stranded DNA binding protein determined by nuclear magnetic resonance spectroscopy.
Nucleic Acids Res., 42, 2014
|
|
6JPW
 
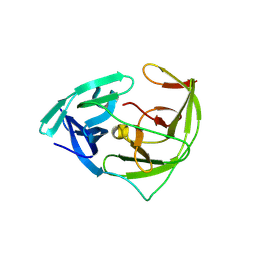 | | Crystal structure of Zika NS2B-NS3 protease with compound 1C | | Descriptor: | NS3 protease, SER-C0F-GLY-LYS-ARG-LYS, Serine protease subunit NS2B | | Authors: | Quek, J.P. | | Deposit date: | 2019-03-28 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Biocompatible Macrocyclization between Cysteine and 2-Cyanopyridine Generates Stable Peptide Inhibitors.
Org.Lett., 21, 2019
|
|
4GX8
 
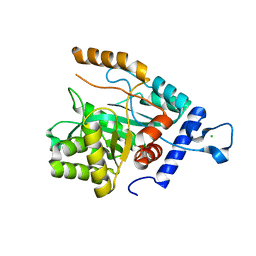 | | Crystal structure of a DNA polymerase III alpha-epsilon chimera | | Descriptor: | CHLORIDE ION, DNA polymerase III subunit epsilon,DNA polymerase III subunit alpha | | Authors: | Robinson, A, Horan, N, Xu, Z.-Q, Dixon, N.E, Oakley, A.J. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits alpha, {varepsilon}, theta and beta reveals a highly flexible arrangement of the proofreading domain
Nucleic Acids Res., 41, 2013
|
|
8VRH
 
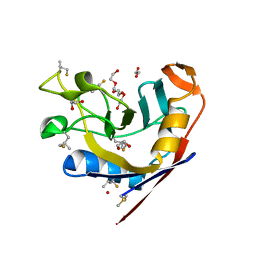 | | E. coli peptidyl-prolyl cis-trans isomerase containing delta2-monofluoro-leucines | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Frkic, R.L, Jackson, C.J. | | Deposit date: | 2024-01-22 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational Preferences of the Non-Canonical Amino Acids (2 S ,4 S )-5-Fluoroleucine, (2 S ,4 R )-5-Fluoroleucine, and 5,5'-Difluoroleucine in a Protein.
Biochemistry, 63, 2024
|
|
