8IN9
 
 | | The structure of the GfsA KSQ-AT didomain in complex with the GfsA ACP domain | | Descriptor: | N-[2-(acetylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, Polyketide synthase | | Authors: | Chisuga, T, Murakami, S, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2023-03-09 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure-Based Analysis of Transient Interactions between Ketosynthase-like Decarboxylase and Acyl Carrier Protein in a Loading Module of Modular Polyketide Synthase.
Acs Chem.Biol., 18, 2023
|
|
3AGV
 
 | | Crystal structure of a human IgG-aptamer complex | | Descriptor: | 5'-R(*GP*GP*AP*GP*GP*(UFT)P*GP*(CFZ)P*(UFT)P*(CFZ)P*(CFZ)P*GP*AP*AP*A*GP*GP*AP*AP*(CFZ)P*(UFT)P*(CFZ)P*(CFZ)P*A)-3', CALCIUM ION, Ig gamma-1 chain C region, ... | | Authors: | Nomura, Y, Sugiyama, S, Sakamoto, T, Miyakawa, S, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Nakamura, Y, Matsumura, H. | | Deposit date: | 2010-04-08 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational plasticity of RNA for target recognition as revealed by the 2.15 A crystal structure of a human IgG-aptamer complex
Nucleic Acids Res., 38, 2010
|
|
2ZUQ
 
 | | Crystal structure of DsbB-Fab complex | | Descriptor: | Disulfide bond formation protein B, Fab fragment heavy chain, Fab fragment light chain, ... | | Authors: | Inaba, K, Suzuki, M, Murakami, S. | | Deposit date: | 2008-10-28 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dynamic nature of disulphide bond formation catalysts revealed by crystal structures of DsbB
Embo J., 28, 2009
|
|
2ZUP
 
 | | Updated crystal structure of DsbB-DsbA complex from E. coli | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein dsbA, UBIQUINONE-1, ... | | Authors: | Inaba, K, Suzuki, M, Murakami, S, Nakagawa, A. | | Deposit date: | 2008-10-28 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Dynamic nature of disulphide bond formation catalysts revealed by crystal structures of DsbB
Embo J., 28, 2009
|
|
2W1B
 
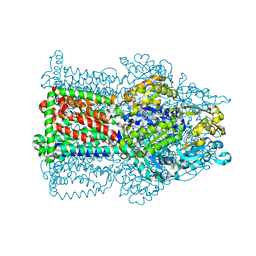 | | The structure of the efflux pump AcrB in complex with bile acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, ACRIFLAVIN RESISTANCE PROTEIN B | | Authors: | Drew, D, Klepsch, M.M, Newstead, S, Flaig, R, De Gier, J.W, Iwata, S, Beis, K. | | Deposit date: | 2008-10-17 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | The Structure of the Efflux Pump Acrb in Complex with Bile Acid.
Mol.Membr.Biol., 25, 2008
|
|
4KXT
 
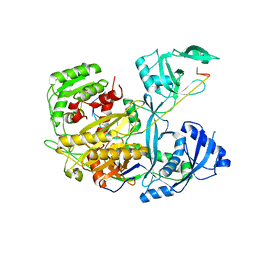 | |
3SUH
 
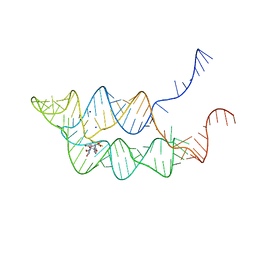 | | Crystal structure of THF riboswitch, bound with 5-formyl-THF | | Descriptor: | N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, Riboswitch, SODIUM ION | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Long-range pseudoknot interactions dictate the regulatory response in the tetrahydrofolate riboswitch.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3SUX
 
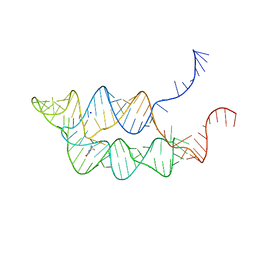 | | Crystal structure of THF riboswitch, bound with THF | | Descriptor: | 5-HYDROXYMETHYLENE-6-HYDROFOLIC ACID, Riboswitch, SODIUM ION | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Long-range pseudoknot interactions dictate the regulatory response in the tetrahydrofolate riboswitch.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3SUY
 
 | |
4WBK
 
 | | The 1.37 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with stearic acid | | Descriptor: | Fatty acid-binding protein, heart, STEARIC ACID | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-09-03 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Molecular Dynamics Simulations of Heart-type Fatty Acid Binding Protein in Apo and Holo Forms, and Hydration Structure Analyses in the Binding Cavity
J.Phys.Chem.B, 119, 2015
|
|
9J4H
 
 | | Crystal structure of SHMT apo form | | Descriptor: | Serine hydroxymethyltransferase, TRIETHYLENE GLYCOL | | Authors: | Murayama, K, Hayashi, H. | | Deposit date: | 2024-08-09 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | SHIN-2 exerts potent activity against VanA-type vancomycin-resistant Enterococcus faecium in vitro by stabilizing the active site loop of serine hydroxymethyltransferase.
Arch.Biochem.Biophys., 761, 2024
|
|
9J4G
 
 | | Crystal structure of SHMT from E. faecium with (+)-SHIN-2 | | Descriptor: | (+)-SHIN-2, Serine hydroxymethyltransferase, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Hayashi, H, Murayama, K. | | Deposit date: | 2024-08-09 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SHIN-2 exerts potent activity against VanA-type vancomycin-resistant Enterococcus faecium in vitro by stabilizing the active site loop of serine hydroxymethyltransferase.
Arch.Biochem.Biophys., 761, 2024
|
|
7PCU
 
 | | Crystal structure of YTHDF1 YTH domain in complex with ebselen | | Descriptor: | N-phenyl-2-selanylbenzamide, YTH domain-containing family protein 1 | | Authors: | Dalle Vedove, A, Cazzanelli, G, Quattrone, A, Provenzani, A, Lolli, G. | | Deposit date: | 2021-08-04 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Small-Molecule Ebselen Binds to YTHDF Proteins Interfering with the Recognition of N 6 -Methyladenosine-Modified RNAs.
Acs Pharmacol Transl Sci, 5, 2022
|
|
7QKN
 
 | | Crystal structure of YTHDF1 YTH domain in complex with the ebsulfur derivative compound 7 | | Descriptor: | 1,2-ETHANEDIOL, THIOCYANATE ION, YTH domain-containing family protein 1, ... | | Authors: | Dalle Vedove, A, Cazzanelli, G, Quattrone, A, Provenzani, A, Lolli, G. | | Deposit date: | 2021-12-18 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Small-Molecule Ebselen Binds to YTHDF Proteins Interfering with the Recognition of N 6 -Methyladenosine-Modified RNAs.
Acs Pharmacol Transl Sci, 5, 2022
|
|
7QL7
 
 | | Crystal structure of YTHDF1 YTH domain in complex with the ebsulfur derivative compound 9 | | Descriptor: | 1,2-ETHANEDIOL, THIOCYANATE ION, YTH domain-containing family protein 1, ... | | Authors: | Dalle Vedove, A, Cazzanelli, G, Quattrone, A, Provenzani, A, Lolli, G. | | Deposit date: | 2021-12-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-Molecule Ebselen Binds to YTHDF Proteins Interfering with the Recognition of N 6 -Methyladenosine-Modified RNAs.
Acs Pharmacol Transl Sci, 5, 2022
|
|
4DOV
 
 | | Structure of free mouse ORC1 BAH domain | | Descriptor: | Origin recognition complex subunit 1 | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2012-02-10 | | Release date: | 2012-03-07 | | Last modified: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome.
Nature, 484, 2012
|
|
4DOW
 
 | | Structure of mouse ORC1 BAH domain bound to H4K20me2 | | Descriptor: | Histone H4, Origin recognition complex subunit 1 | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2012-02-10 | | Release date: | 2012-03-07 | | Last modified: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome.
Nature, 484, 2012
|
|
4DA4
 
 | |
8C58
 
 | | CpG specific M.MpeI methyltransferase crystallized in the presence of 5-hydroxycytosine and 5-methylcytosine containing dsDNA | | Descriptor: | CARBONATE ION, Cytosine-specific methyltransferase, DNA (5'-D(*CP*CP*AP*CP*AP*TP*GP*(5OC)P*GP*CP*TP*GP*AP*A)-3'), ... | | Authors: | Wojciechowski, M, Czapinska, H, Krwawicz, J, Rafalski, D, Bochtler, M. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cytosine analogues as DNA methyltransferase substrates.
Nucleic Acids Res., 52, 2024
|
|
8C59
 
 | | CpG specific M.MpeI methyltransferase crystallized in the presence of 5-bromocytosine (converted to 5mC) and 5-methylcytosine containing dsDNA | | Descriptor: | CARBONATE ION, CITRIC ACID, Cytosine-specific methyltransferase, ... | | Authors: | Wojciechowski, M, Czapinska, H, Krwawicz, J, Rafalski, D, Bochtler, M. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cytosine analogues as DNA methyltransferase substrates.
Nucleic Acids Res., 52, 2024
|
|
8C57
 
 | | CpG specific M.MpeI methyltransferase crystallized in the presence of 5,6-dihydro-5-azacytosine (converted to 5m-dhaC) and 5-methylcytosine containing dsDNA | | Descriptor: | CARBONATE ION, Cytosine-specific methyltransferase, DNA (5'-D(*CP*CP*AP*CP*AP*TP*GP*(5MA)P*GP*CP*TP*GP*AP*A)-3'), ... | | Authors: | Wojciechowski, M, Czapinska, H, Krwawicz, J, Rafalski, D, Bochtler, M. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cytosine analogues as DNA methyltransferase substrates.
Nucleic Acids Res., 52, 2024
|
|
8C56
 
 | | CpG specific M.MpeI methyltransferase crystallized in the presence of 2'-deoxy-5-methylzebularine (5mZ) and 5-methylcytosine containing dsDNA | | Descriptor: | Cytosine-specific methyltransferase, DNA (5'-D(*CP*CP*AP*CP*AP*TP*GP*(5PY)P*GP*CP*TP*GP*AP*A)-3'), DNA (5'-D(*GP*TP*TP*CP*AP*GP*(5CM)P*GP*CP*AP*TP*GP*TP*G)-3'), ... | | Authors: | Wojciechowski, M, Czapinska, H, Krwawicz, J, Rafalski, D, Bochtler, M. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cytosine analogues as DNA methyltransferase substrates.
Nucleic Acids Res., 52, 2024
|
|
3AQO
 
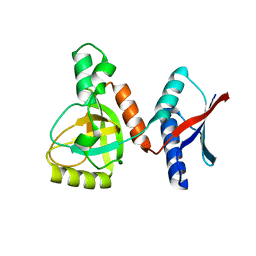 | |
