2VBJ
 
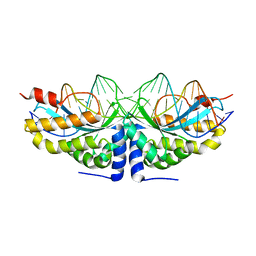 | | Molecular basis of human XPC gene recognition and cleavage by engineered homing endonuclease heterodimers | | Descriptor: | 5'-D(*TP*CP*TP*GP*CP*CP*TP*TP*TP*TP *TP*TP*GP*AP*AP*GP*GP*AP*TP*CP*CP*TP*AP*A)-3', 5'-D(*TP*TP*AP*GP*GP*AP*TP*CP*CP*TP *TP*CP*AP*AP*AP*AP*AP*AP*GP*GP*CP*AP*GP*A)-3', CALCIUM ION, ... | | Authors: | Redondo, P, Prieto, J, Munoz, I.G, Alibes, A, Stricher, F, Serrano, L, Arnould, S, Perez, C, Cabaniols, J.P, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2007-09-14 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Basis of Xeroderma Pigmentosum Group C DNA Recognition by Engineered Meganucleases
Nature, 456, 2008
|
|
2VBO
 
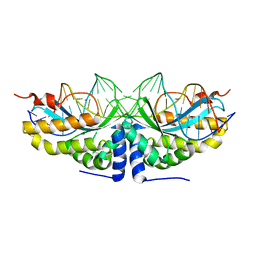 | | Molecular basis of human XPC gene recognition and cleavage by engineered homing endonuclease heterodimers | | Descriptor: | 5'-D(*TP*CP*TP*GP*CP*CP*TP*TP*TP*TP *TP*TP*GP*AP*AP*GP*GP*AP*TP*CP*CP*TP*AP*A)-3', 5'-D(*TP*TP*AP*GP*GP*AP*TP*CP*CP*TP *TP*CP*AP*AP*AP*AP*AP*AP*GP*GP*CP*AP*GP*A)-3', CALCIUM ION, ... | | Authors: | Redondo, P, Prieto, J, Munoz, I.G, Alibes, A, Stricher, F, Serrano, L, Arnould, S, Perez, C, Cabaniols, J.P, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2007-09-14 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis of Xeroderma Pigmentosum Group C DNA Recognition by Engineered Meganucleases
Nature, 456, 2008
|
|
7ODF
 
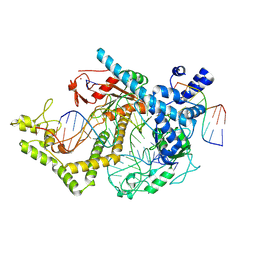 | | Structure of the mini-RNA-guided endonuclease CRISPR-Cas_phi3 | | Descriptor: | Cas_phi3, DNA (5'-D(P*GP*G)-3'), DNA (5'-D(P*GP*TP*AP*AP*TP*TP*CP*AP*G)-3'), ... | | Authors: | Carabias del Rey, A, Fugilsang, A, Temperini, P, Pape, T, Sofos, N, Stella, S, Erledsson, S, Montoya, G. | | Deposit date: | 2021-04-29 | | Release date: | 2021-07-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structure of the mini-RNA-guided endonuclease CRISPR-Cas12j3.
Nat Commun, 12, 2021
|
|
8RWG
 
 | | PAO1 wild-type ribosome, R, reference map | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mesa, P, Montoya, G. | | Deposit date: | 2024-02-04 | | Release date: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Detuning of the Ribosome Conformational Landscape Promotes Antibiotic Resistance and Collateral Sensitivity.
To Be Published
|
|
2YPF
 
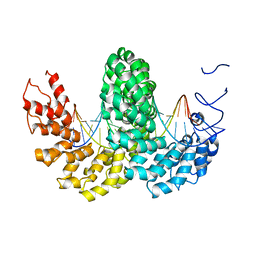 | | Structure of the AvrBs3-DNA complex provides new insights into the initial thymine-recognition mechanism | | Descriptor: | 5'-D(*TP*AP*GP*AP*GP*GP*GP*TP*TP*AP*GP*GP*TP*TP *TP*AP*TP*AP*TP*AP*AP)-3', 5'-D(*TP*TP*TP*AP*TP*AP*TP*AP*AP*AP*CP*CP*TP*AP *AP*CP*CP*CP*TP*CP*TP*AP)-3', AVRBS3 | | Authors: | Stella, S, Molina, R, Yefimenko, I, Prieto, J, Silva, G.H, Bertonati, C, Juillerat, A, Duchateau, P, Montoya, G. | | Deposit date: | 2012-10-30 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the Avrbs3-DNA Complex Provides New Insights Into the Initial Thymine-Recognition Mechanism
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8EXA
 
 | | ISDra2 TnpB in complex with reRNA and cognate DNA, conformation 1 (RuvC domain resolved) | | Descriptor: | DNA (43-MER), RNA (150-MER), RNA-guided DNA endonuclease TnpB | | Authors: | Sasnauskas, G, Tamulaitiene, G, Carabias, A, Karvelis, T, Druteika, G, Silanskas, A, Montoya, G, Venclovas, C, Kazlauskas, D, Siksnys, V. | | Deposit date: | 2022-10-25 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | TnpB structure reveals minimal functional core of Cas12 nuclease family.
Nature, 616, 2023
|
|
8EX9
 
 | | ISDra2 TnpB in complex with reRNA and cognate DNA, conformation 2 (RuvC domain unresolved) | | Descriptor: | DNA (43-MER), RNA (150-MER), RNA-guided DNA endonuclease TnpB | | Authors: | Sasnauskas, G, Tamulaitiene, G, Carabias, A, Karvelis, T, Druteika, G, Silanskas, A, Montoya, G, Venclovas, C, Kazlauskas, D, Siksnys, V. | | Deposit date: | 2022-10-25 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | TnpB structure reveals minimal functional core of Cas12 nuclease family.
Nature, 616, 2023
|
|
7Z38
 
 | | Structure of the RAF1-HSP90-CDC37 complex (RHC-I) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37, ... | | Authors: | Mesa, P, Garcia-Alonso, S, Barbacid, M, Montoya, G. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure of the RAF1-HSP90-CDC37 complex reveals the basis of RAF1 regulation.
Mol.Cell, 82, 2022
|
|
7Z37
 
 | | Structure of the RAF1-HSP90-CDC37 complex (RHC-II) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37, ... | | Authors: | Mesa, P, Garcia-Alonso, S, Barbacid, M, Montoya, G. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structure of the RAF1-HSP90-CDC37 complex reveals the basis of RAF1 regulation.
Mol.Cell, 82, 2022
|
|
2OGQ
 
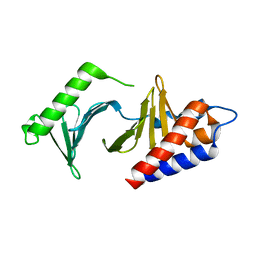 | | Molecular and structural basis of Plk1 substrate recognition: Implications in centrosomal localization | | Descriptor: | Serine/threonine-protein kinase PLK1 | | Authors: | Garcia-Alvarez, B, de Carcer, G, Ibanez, S, Bragado-Nilsson, E, Montoya, G. | | Deposit date: | 2007-01-08 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular and structural basis of polo-like kinase 1 substrate recognition: Implications in centrosomal localization.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OJX
 
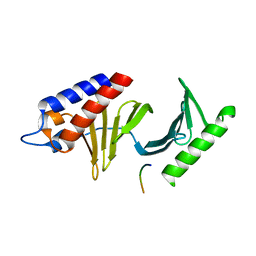 | | Molecular and structural basis of polo-like kinase 1 substrate recognition: Implications in centrosomal localization | | Descriptor: | Serine/threonine-protein kinase PLK1, Synthetic peptide | | Authors: | Garcia-Alvarez, B, de Carcer, G, Ibanez, S, Bragado-Nilsson, E, Montoya, G. | | Deposit date: | 2007-01-15 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular and structural basis of polo-like kinase 1 substrate recognition: Implications in centrosomal localization.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4UN8
 
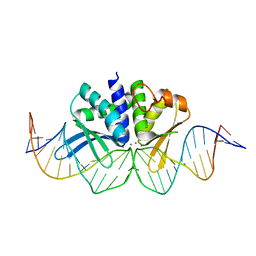 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 1H INCUBATION IN 5MM MN (STATE 2) | | Descriptor: | 25MER, HOMING ENDONUCLEASE I-DMOI, MANGANESE (II) ION | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UNA
 
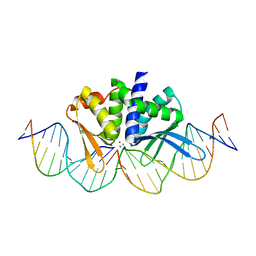 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 2 DAYS INCUBATION IN 5MM MN (STATE 4) | | Descriptor: | 25MER, 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*C)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*C)-3', ... | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UN9
 
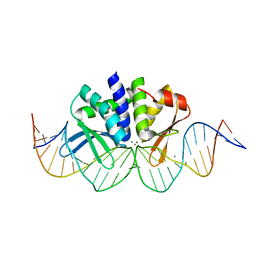 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 8H INCUBATION IN 5MM MN (STATE 3) | | Descriptor: | 25MER, HOMING ENDONUCLEASE I-DMOI, MANGANESE (II) ION | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UNC
 
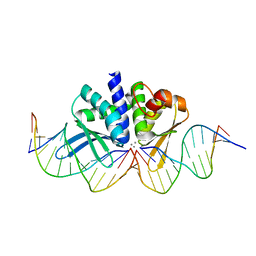 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 8 DAYS INCUBATION IN 5MM MN (STATE 6) | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*C)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*C)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*A)-3', ... | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UNB
 
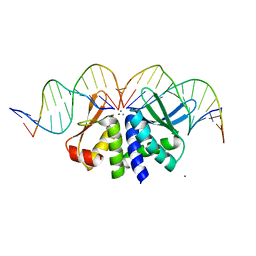 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 6 DAYS INCUBATION IN 5MM MN (STATE 5) | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*C)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*C)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*A)-3', ... | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UN7
 
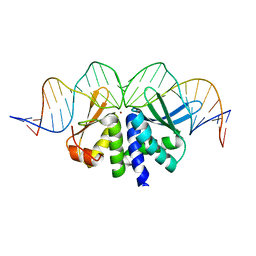 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA BEFORE INCUBATION IN 5MM MN (STATE 1) | | Descriptor: | 25MER, HOMING ENDONUCLEASE I-DMOI, ZINC ION | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
2VTX
 
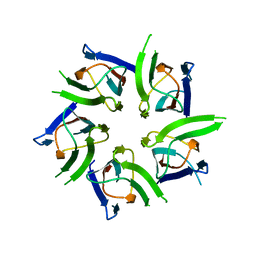 | | ACTIVATION OF NUCLEOPLASMIN, AN OLIGOMERIC HISTONE CHAPERONE, CHALLENGES ITS STABILITY | | Descriptor: | NPM-A PROTEIN | | Authors: | Taneva, S.G, Munoz, I.G, Franco, G, Falces, J, Arregi, I, Muga, A, Montoya, G, Urbaneja, M.A, Banuelos, S. | | Deposit date: | 2008-05-16 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Activation of Nucleoplasmin, an Oligomeric Histone Chaperone, Challenges its Stability.
Biochemistry, 47, 2008
|
|
1QZX
 
 | |
7Z55
 
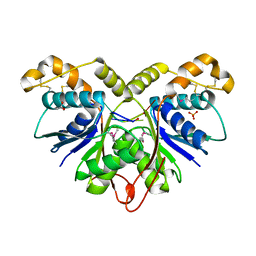 | | Crystal Structure of the Ring Nuclease 0455 from Sulfolobus islandicus (Sis0455) in complex with its substrate | | Descriptor: | ACETATE ION, CRISPR-associated protein, Cyclic RNA cA4, ... | | Authors: | Molina, R, Martin-Garcia, R, Lopez-Mendez, B, Jensen, A.L.G, Marchena-Hurtado, J, Stella, S, Montoya, G. | | Deposit date: | 2022-03-08 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.664 Å) | | Cite: | Molecular basis of cyclic tetra-oligoadenylate processing by small standalone CRISPR-Cas ring nucleases.
Nucleic Acids Res., 50, 2022
|
|
7Z56
 
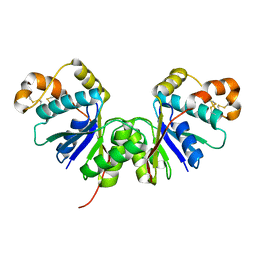 | | Crystal Structure of the Ring Nuclease 0455 from Sulfolobus islandicus (Sis0455) in its apo form | | Descriptor: | CRISPR-associated protein | | Authors: | Molina, R, Martin-Garcia, R, Lopez-Mendez, B, Jensen, A.L.G, Marchena-Hurtado, J, Stella, S, Montoya, G. | | Deposit date: | 2022-03-08 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Molecular basis of cyclic tetra-oligoadenylate processing by small standalone CRISPR-Cas ring nucleases.
Nucleic Acids Res., 50, 2022
|
|
2XSM
 
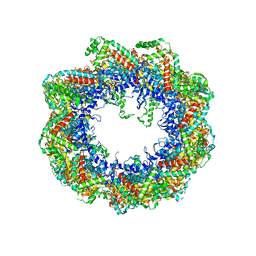 | | Crystal structure of the mammalian cytosolic chaperonin CCT in complex with tubulin | | Descriptor: | CCT | | Authors: | Munoz, I.G, Yebenes, H, Zhou, M, Mesa, P, Serna, M, Bragado-Nilsson, E, Beloso, A, Robinson, C.V, Valpuesta, J.M, Montoya, G. | | Deposit date: | 2010-10-29 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Crystal Structure of the Open Conformation of the Mammalian Chaperonin Cct in Complex with Tubulin.
Nat.Struct.Mol.Biol., 18, 2011
|
|
8AA5
 
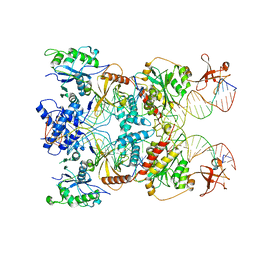 | | Cryo-EM structure of the strand transfer complex of the TnsB transposase (type V-K CRISPR-associated transposon) | | Descriptor: | LE_PolyA, LE_Target, RE_PolyA, ... | | Authors: | Tenjo-Castano, F, Sofos, N, Lopez-Mendez, B, Stutzke, L.S, Fuglsang, A, Stella, S, Montoya, G. | | Deposit date: | 2022-06-30 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structure of the TnsB transposase-DNA complex of type V-K CRISPR-associated transposon.
Nat Commun, 13, 2022
|
|
1QZW
 
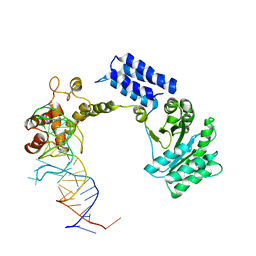 | | Crystal structure of the complete core of archaeal SRP and implications for inter-domain communication | | Descriptor: | 7S RNA, Signal recognition 54 kDa protein | | Authors: | Rosendal, K.R, Wild, K, Montoya, G, Sinning, I. | | Deposit date: | 2003-09-18 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of the complete core of archaeal signal recognition particle and implications for interdomain communication
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2JA4
 
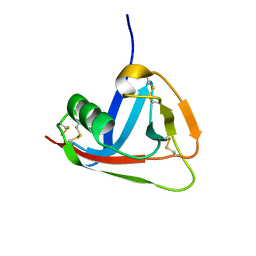 | | Crystal structure of CD5 domain III reveals the fold of a group B scavenger cysteine-rich receptor | | Descriptor: | T-CELL SURFACE GLYCOPROTEIN CD5 | | Authors: | Rodamilans, B, Munoz, I.G, Sarrias, M.R, Lozano, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2006-11-21 | | Release date: | 2007-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of the Third Extracellular Domain of Cd5 Reveals the Fold of a Group B Scavenger Cysteine-Rich Receptor Domain.
J.Biol.Chem., 282, 2007
|
|
