1UYW
 
 | | Crystal Structure of the antiflavivirus Fab4g2 | | Descriptor: | FAB ANTIBODY HEAVY CHAIN, FAB ANTIBODY LIGHT CHAIN | | Authors: | Martinez-Fleites, C, Ortiz-Lombardia, M, Taylor, E.J, Gil-Valdes, J, Chinea, G, Davies, G. | | Deposit date: | 2004-03-03 | | Release date: | 2005-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Antiflavivirus Fab4G2
To be Published
|
|
4DTH
 
 | | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with ATP and Mg++ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Durand, E, Audoly, G, Derrez, E, Spinelli, S, Ortiz-Lombardia, M, Cascales, E, Raoult, D, Cambillau, C. | | Deposit date: | 2012-02-21 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with ATP and Mg++
J.Biol.Chem., 2012
|
|
4DTD
 
 | | Structure and functional characterization of a Vibrio cholerae toxin from the MARTX/VgrG family. | | Descriptor: | GLYCEROL, VgrG protein | | Authors: | Durand, E, Audoly, G, Derrez, E, Spinelli, S, Ortiz-Lombardia, M, Cascales, E, Raoult, D, Cambillau, C. | | Deposit date: | 2012-02-21 | | Release date: | 2012-08-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and functional characterization of a Vibrio cholerae toxin
from the MARTX/VgrG family.
J.Biol.Chem., 2012
|
|
4DTL
 
 | | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with ATP and Mn++ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Durand, E, Audoly, G, Derrez, E, Spinelli, S, Ortiz-Lombardia, M, Cascales, E, Raoult, D, Cambillau, C. | | Deposit date: | 2012-02-21 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with ATP and Mn++
J.Biol.Chem., 2012
|
|
4E1F
 
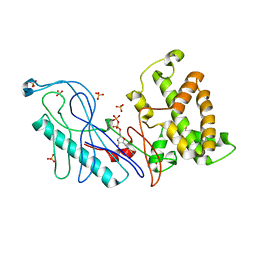 | | Structure of a VgrG Vibrio cholerae toxin ACD domain Glu16Gln mutant in complex with ADP and Mn++ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Durand, E, Audoly, G, Derrez, E, Spinelli, S, Ortiz-Lombardia, M, Cascales, E, Raoult, D, Cambillau, C. | | Deposit date: | 2012-03-06 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and functional characterization of the Vibrio cholerae toxin
from the VgrG/MARTX family.
J.Biol.Chem., 2012
|
|
2C5S
 
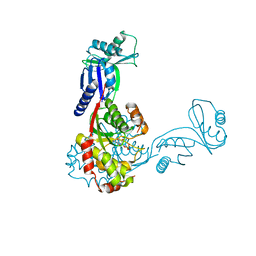 | | Crystal structure of Bacillus anthracis ThiI, a tRNA-modifying enzyme containing the predicted RNA-binding THUMP domain | | Descriptor: | ADENOSINE MONOPHOSPHATE, PROBABLE THIAMINE BIOSYNTHESIS PROTEIN THII | | Authors: | Waterman, D.G, Ortiz-Lombardia, M, Fogg, M.J, Koonin, E.V, Antson, A.A. | | Deposit date: | 2005-11-01 | | Release date: | 2005-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Bacillus anthracis ThiI, a tRNA-modifying enzyme containing the predicted RNA-binding THUMP domain.
J.Mol.Biol., 356, 2006
|
|
3RX9
 
 | | 3D structure of SciN from an Escherichia coli Patotype | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Felisberto-Rodrigues, C, Durand, E, Aschtgen, M.-S, Blangy, S, Ortiz-Lombardia, M, Douzy, B, Cambillau, C, Cascales, E. | | Deposit date: | 2011-05-10 | | Release date: | 2011-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Towards a Structural Comprehension of Bacterial Type VI Secretion Systems: Characterization of the TssJ-TssM Complex of an Escherichia coli Pathovar.
Plos Pathog., 7, 2011
|
|
2X53
 
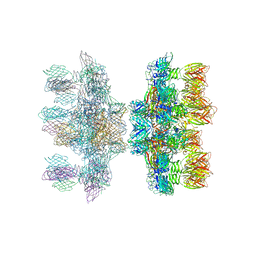 | | Structure of the phage p2 baseplate in its activated conformation with Sr | | Descriptor: | ORF15, ORF16, PUTATIVE RECEPTOR BINDING PROTEIN, ... | | Authors: | Sciara, G, Bebeacua, C, Bron, P, Tremblay, D, Ortiz-Lombardia, M, Lichiere, J, van Heel, M, Campanacci, V, Moineau, S, Cambillau, C. | | Deposit date: | 2010-02-05 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of Lactococcal Phage P2 Baseplate and its Mechanism of Activation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XYQ
 
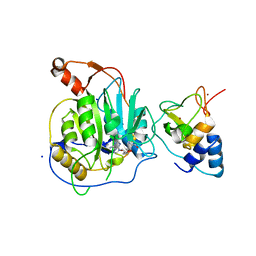 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
2WZP
 
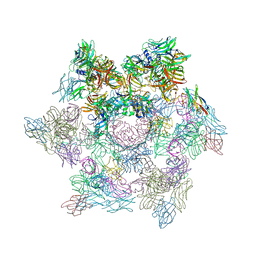 | | Structures of Lactococcal Phage p2 Baseplate Shed Light on a Novel Mechanism of Host Attachment and Activation in Siphoviridae | | Descriptor: | CAMELID VHH5, LACTOCOCCAL PHAGE P2 ORF15, LACTOCOCCAL PHAGE P2 ORF16, ... | | Authors: | Sciara, G, Bebeacua, C, Bron, P, Tremblay, D, Ortiz-Lombardia, M, Lichiere, J, van Heel, M, Campanacci, V, Moineau, S, Cambillau, C. | | Deposit date: | 2009-12-01 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Lactococcal Phage P2 Baseplate and its Mechanism of Activation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XUS
 
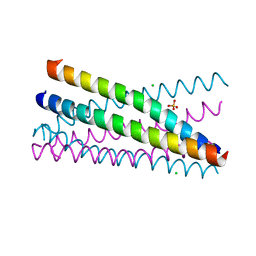 | | Crystal Structure of the BRMS1 N-terminal region | | Descriptor: | BREAST CANCER METASTASIS-SUPPRESSOR 1, CHLORIDE ION, SULFATE ION | | Authors: | Spinola-Amilibia, M, Rivera, J, Ortiz-Lombardia, M, Romero, A, Neira, J.L, Bravo, J. | | Deposit date: | 2010-10-20 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | The Structure of Brms1 Nuclear Export Signal and Snx6 Interacting Region Reveals a Hexamer Formed by Antiparallel Coiled Coils.
J.Mol.Biol., 411, 2011
|
|
2XYR
 
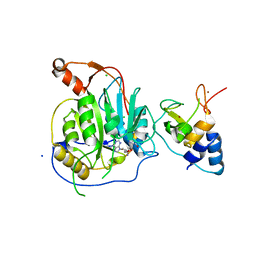 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
2XYV
 
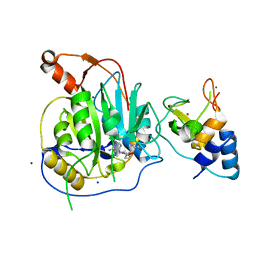 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-19 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
3ZQQ
 
 | | Crystal structure of the full-length small terminase from a SPP1-like bacteriophage | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZQM
 
 | | Crystal structure of the small terminase oligomerization core domain from a SPP1-like bacteriophage (crystal form 1) | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2WKC
 
 | |
467D
 
 | |
3ZQO
 
 | | Crystal structure of the small terminase oligomerization core domain from a SPP1-like bacteriophage (crystal form 3) | | Descriptor: | POTASSIUM ION, TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZQP
 
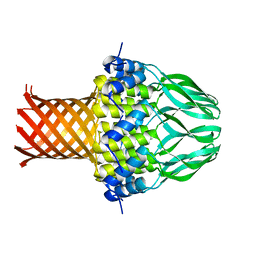 | | Crystal structure of the small terminase oligomerization domain from a SPP1-like bacteriophage | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZQN
 
 | | Crystal structure of the small terminase oligomerization core domain from a SPP1-like bacteriophage (crystal form 2) | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Buttner, C.R, Chechik, M, Ortiz-Lombardia, M, Smits, C, Chechik, V, Jeschke, G, Dykeman, E, Benini, S, Alonso, J.C, Antson, A.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for DNA Recognition and Loading Into a Viral Packaging Motor.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2WKD
 
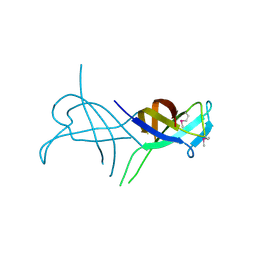 | |
2WBM
 
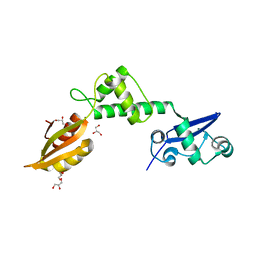 | | Crystal structure of mthSBDS, the homologue of the Shwachman-Bodian- Diamond syndrome protein in the euriarchaeon Methanothermobacter thermautotrophicus | | Descriptor: | CHLORIDE ION, GLYCEROL, RIBOSOME MATURATION PROTEIN SDO1 HOMOLOG, ... | | Authors: | Ng, C.L, Isupov, M.N, Lebedev, A.A, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2009-03-02 | | Release date: | 2009-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conformational Flexibility and Molecular Interactions of an Archaeal Homologue of the Shwachman-Bodian-Diamond Syndrome Protein.
Bmc Struct.Biol., 9, 2009
|
|
2WNR
 
 | | The structure of Methanothermobacter thermautotrophicus exosome core assembly | | Descriptor: | PHOSPHATE ION, PROBABLE EXOSOME COMPLEX EXONUCLEASE 1, PROBABLE EXOSOME COMPLEX EXONUCLEASE 2 | | Authors: | Ng, C.L, Waterman, D.G, Antson, A.A, Ortiz-Lombardia, M. | | Deposit date: | 2009-07-19 | | Release date: | 2010-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the Methanothermobacter Thermautotrophicus Exosome Rnase Ph Ring
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2XF7
 
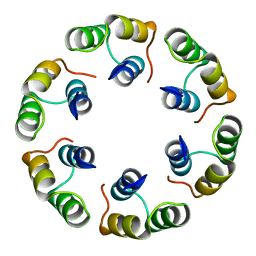 | | Crystal structure of Bacillus subtilis SPP1 phage gp23.1, a putative chaperone. High-resolution structure. | | Descriptor: | GP23.1 | | Authors: | Veesler, D, Blangy, S, Lichiere, J, Ortiz-Lombardia, M, Tavares, P, Campanacci, V, Cambillau, C. | | Deposit date: | 2010-05-20 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of Bacillus Subtilis Spp1 Phage Gp23.1, A Putative Chaperone.
Protein Sci., 19, 2010
|
|
2WNY
 
 | | Structure of Mth689, a DUF54 protein from Methanothermobacter thermautotrophicus | | Descriptor: | CHLORIDE ION, CONSERVED PROTEIN MTH689, NICKEL (II) ION | | Authors: | Ng, C.L, Waterman, D.G, Lebedev, A.A, Smits, C, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2009-07-21 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Mth689, a Duf54 Protein from Methanothermobacter Thermautotrophicus
To be Published
|
|
